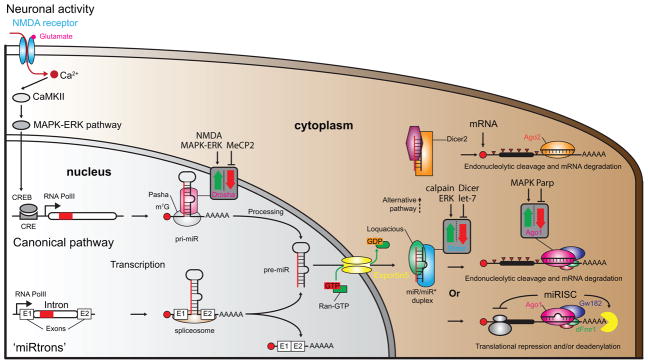
Figure 1. Biogenesis of microRNAs.
MiR gene expression is regulated in ways similar to protein coding genes. Neuronal activity (depolarization, neurotrophins, sensory stimuli, etc.) can induce miR expression [12,17,82]. MiR-132, for example, possesses a CRE sequence in its promoter allowing CREB to regulate its activity. The NMDA receptor, CaMKII and the MAPK-ERK pathway are known to control miR-132 expression although different upstream signaling pathways that may regulate other miR genes [12]. There are two miR biogenesis pathways in the nucleus, the canonical and the ‘miRtrons’ pathway. In the canonical pathway, RNA polymerase II transcribes miR genes into pri-miRs that are further processed into pre-miRs by the Pasha/Drosha microprocessor complex. In the miRtron pathway, miRs are spliced from introns by the spliceosome to pre-miRs. Exportin5 along with Ran-GTP actively transports pre-miRs to the cytoplasm. Dicer/Loquacious complex further processes the pre-miRs in the cytoplasm to mature duplexes - with a guide strand (miR) and its passenger (miR*). A member of the Argonaute family of proteins (Ago1 or Ago2 depending on the pathway) loads one of the strands into the miRISC complex (lower right corner). The complex includes the Gw182 protein and other auxiliary proteins such as dFmr1. This outcome leads to mRNA degradation or translational inhibition depending on the match quality of miR/mRNA hybrid. In rare cases (upper right corner), duplexes with a very high level of complementarity between strands are sorted to an alternative pathway using Dicer2, leading to mRNA degradation by Ago2 and inducing competition between the processing pathways [83]. Neural activity, genetic manipulations or disorders have been shown to positively or negatively influence the processing enzymes involved in miRNA biogenesis (grey squares with red and green arrows) [12,82,84].