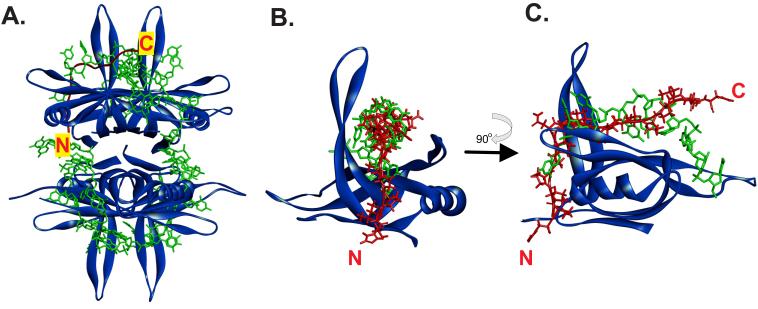
Figure 5. The PXXP motifs of the IDL can bind to the OB-fold in the same position as ssDNA.
(A). Position of a PXXP ligand (i.e., the IDL) bound to one subunit of the SSB tetramer. (B) and (C). The ligand and ssDNA may bind to the same position in the OB-fold. In each panel, SSB is represented as a ribbon diagram and is coloured blue. ssDNA is shown as a stick model in green and the PXXP ligand is coloured in red. In panel A, the ligand is represented as a schematic and in panels B and C as a stick model. The positions of the N- and C-termini of the ligand are indicated. The image in C is rotated 90° relative to that in B as shown by the arrow. The PDB files used in this analysis are 1EYG for SSB and 2KXC for the ligand (Aitio et al., 2010; Raghunathan et al., 2000). The 2KXC structure was used in this analysis as the ligand contains tandem PXXP motifs similar to that in the SSB IDL. To generate the images shown, 2KXC was aligned onto file 1EYG using TM-align (Zhang and Skolnick, 2005). Then, the SH3 domain was removed so that clear visualization of the ligand (IDL) bound to the OB-fold in one monomer can be made.