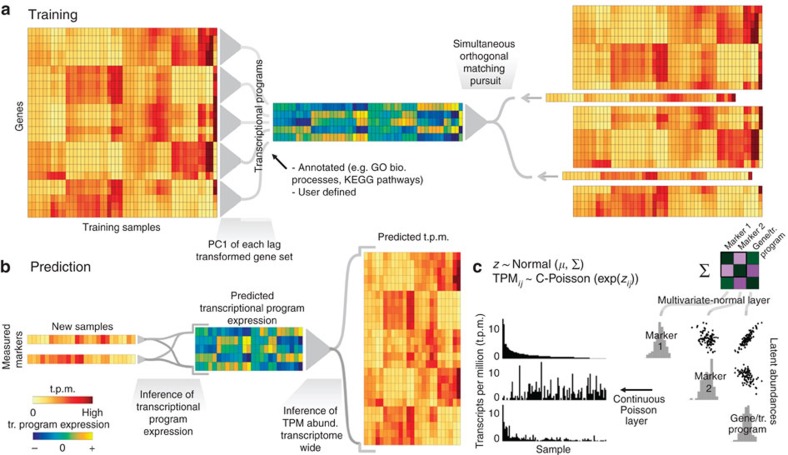
Figure 2. Tradict's algorithmic workflow.
(a) During training, the transcriptome is first quantitatively summarized in terms of a collection of a few hundred, biologically comprehensive transcriptional programs. These are then decomposed into a subset of marker genes using an adaptation of the Simultaneous Orthogonal Matching Pursuit algorithm. A MVN-CP hierarchical model is used as a predictive model to capture covariance relationships between markers, transcriptional programs and all genes. (b) During prediction, Tradict predicts the expression of transcriptional programs and all genes in the transcriptome using the expression measurements of the marker genes. (c) The MVN-CP hierarchy enables Tradict to efficiently model statistical coupling between the non-negative expression measurements typical of sequencing experiments. This is done by assuming that associated with each observed, noisy t.p.m. measurement, there is an unmeasured (denoised), latent abundance the logarithm of which comes from a MVN distribution over all genes and transcriptional programs.