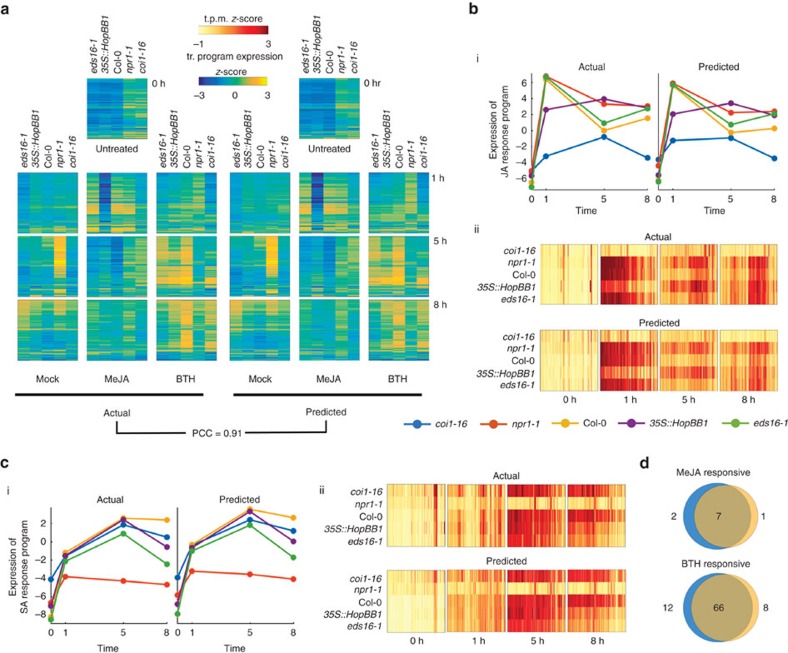
Figure 4. Tradict accurately predicts transcriptional responses across time in response to hormone perturbation in an A. thaliana innate immune signaling data set.
After being trained on the full A. thaliana training transcriptome collection, the selected set of 100 globally representative and context-independent markers were used to predict the expression of transcriptional programs and all genes for the transcriptomes presented in Yang et al.31. (a) Actual versus predicted heatmaps for the expression of all 150 transcriptional programs in A. thaliana across genotype, time and hormone treatment. (b) Predicted versus actual expression of (i) the JA response transcriptional program, and (ii) the genes involved in the JA response program. (c) (i–ii) Same as b, but for the SA response transcriptional program. (d) Hypothesis free, differential transcriptional program expression analysis as performed on the actual expression of transcriptional programs versus those predicted by Tradict. Blue circles represent the actual and orange represent the predicted. All heatmaps are clustered in the same order across time, treatment, genotype and between predicted and actual.