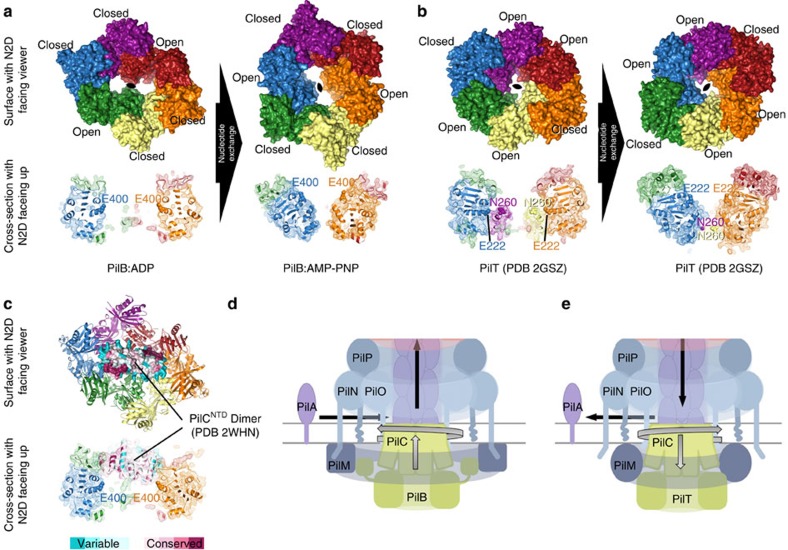
Figure 5. Modeling the movements and functions of PilB and PilT ATPases.
(a) The PilB:ADP hexamer was aligning with the PilB:AMP-PNP hexamer to suggest the movements that may occur as two closed packing units open and two open packing units close during nucleotide exchange. Chains are coloured as in Fig. 2. (Bottom) A cross-section through the centre of PilB is shown with E400 displayed as spheres. (b) The PilT hexamer (PDB 2GSZ) was aligned with the same PilT hexamer rotated by one packing unit to suggest the movements that may occur as two closed packing units open and two open packing units close during nucleotide exchange. (Bottom) A cross-section through the centre of PilT is shown with E222 (the equivalent of E400 in PilB) and N260 from the midpoint of the extension on pore loop 3 (there is no equivalent in PilB) displayed as spheres. (c) The PilCNTD dimer (PDB 2WHN) was manually placed in the two sub-pores of PilB, and the phylogenetic conservation of PilC residues were mapped onto the surface using the ConSurf server51. (Bottom) A cross-section through the centre of PilB is shown with E400 displayed as spheres. The model is similar to the mode of PilC binding to PilB we proposed previously8. (d) Working model for the molecular mechanism of the PilB motor. We propose that PilC is thrusted by PilB upwards towards the membrane, allowed to fall back, and rotated in 60° increments to facilitate helical PilA polymerization. (e) Working model for the molecular mechanism of the PilT motor. We propose that PilC is wrenched downward by PilT towards the cytoplasm, allowed to relax upwards, and rotated in 60° increments in the opposite direction of PilB to facilitate helical PilA depolymerization. The model presented in panels D and E refines our previous model that described how PilM-PilN interactions modulate PilB/PilT associations27.