Figure 2.
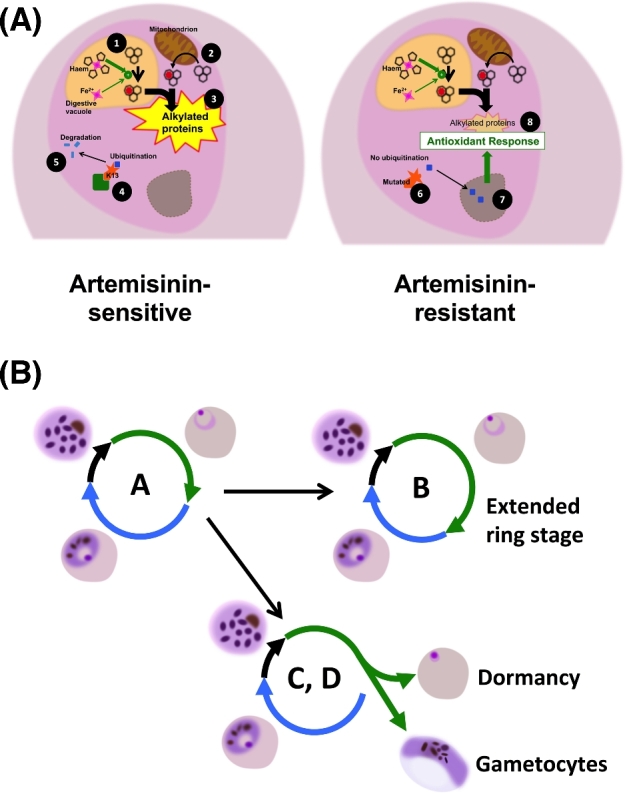
Proposed mechanisms of artemisinin resistance. (A) Relevant biochemical pathways. In ring-stage parasites, artemisinin is primarily activated by haem produced in the process of haemoglobin digestion (1) although haem biosynthesis in mitochondria may also contribute (2). Activated artemisinins alkylate nearby proteins in an indiscriminate manner leading to cell death (3). In artemisinin-sensitive parasites, a transcriptional factor with potential to upregulate protein turnover and oxidative damage responses is bound via the K13 adaptor (4) leading to its ubiquitination and proteolysis (5). K13 mutation disrupts this binding (6) allowing the factor to enter the nucleus (7) with upregulation of a range of transcriptional responses that can mitigate the downstream consequences of artemisinins (8). (B) Proposed phenotypes associated with artemisinin resistance. The overall length and proportion of time spent in each stage appears relatively fixed in a given strain (A). By extending their ring-stage (B), parasites increase the period of reduced vulnerability to artemisinins. An alternative is to increase the proportion of parasites entering dormancy (C), a natural phenomenon observed in all parasite strains that allows escape from relatively short duration artemisinin exposures in patient treatments. Finally, increasing the proportion of parasites that differentiate into gametocytes (D) at a given timepoint could improve chances of transmission before treatment is administered.
