Figure 3. The catalytic subunit does not make stable contact with nucleosomal DNA in the absence of the Snf5 module, but does with the histone octamer surface.
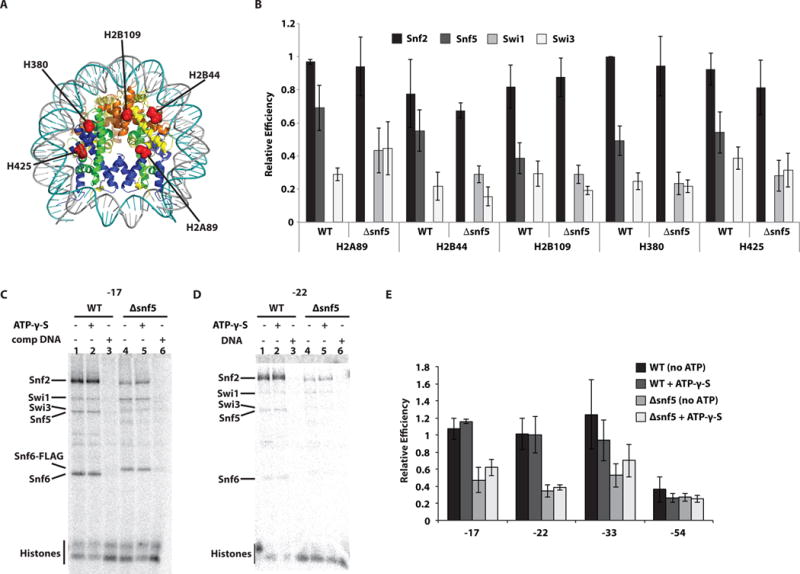
(A) The interactions of wild type (WT) and Δsnf5 SWI/SNF with nucleosomes were mapped by histone photocrosslinking with photoreactive radio-iodinated N-((2-pyridyldithio)ethyl)-4-azidosalicylamide (PEAS) attached to specific histone positions as indicated by histone type and residue position. (B) The relative crosslinking efficiency of Snf2, Snf5, Swi3 and in some instances Swi1 was plotted for each of the five histone positions and for WT and Δsnf5 SWI/SNF. Signals were normalized relative to crosslinking in WT SWI/SNF to residue 80 of histone H3 (H380). (C–D) Purified WT SWI/SNF complex was bound to nucleosomes reconstituted with radiolabeled DNA probes containing photoreactive groups at the positions indicated (“0” corresponding to the dyad). Reactions contained either wild type (lanes 1–3) or Δsnf5 (lanes 4–6) complexes. In some instances 150 μM γ-thio-ATP (lanes 2 and 5) or 13 μg/ml competitor DNA (lanes 3 and 6) were added. (E) The efficiency of Snf2 crosslinking to DNA was plotted for four different positions within nucleosomal DNA and were normalized to nt-17. These experiments were done in triplicates and the standard deviation (SD) is shown.
