Figure 6. Loss of the Snf5 subunit adversely affects the intrinsic nucleosome mobilizing activity of SWI/SNF.
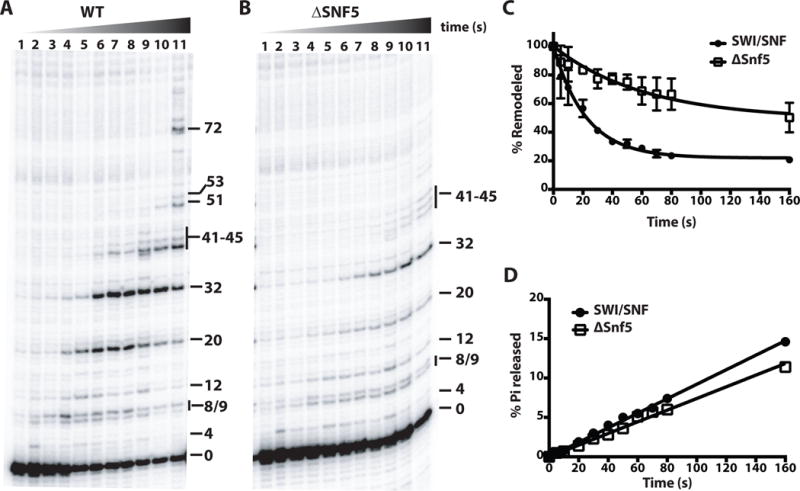
(A–B) Nucleosomes modified at residue 53 of histone H2B were used for monitoring movement of DNA on the octamer surface. DNA cleavage products were resolved on a denaturing 6% polyacrylamide gel and visualized by phosphorimaging. Numbers on the right side of the gel image refer to number of nucleotides (nt) moved from the starting cleavage position (0). Nucleosomes were remodeled with WT (A), and Δsnf5 SWI/SNF (B) for 0, 5, 10, 20, 30, 40, 50, 60, 70, 80 and 160 s using 4.4 μM ATP. (C) The amount of DNA cleaved at starting position (0) was plotted versus time for WT SWI/SNF and Δsnf5 SWI/SNF. Rate constants (k) obtained by fitting data to single exponential function were 0.044± 0.004 s−1 (WT) and 0.016 ± 0.005 s−1 (Δsnf5). Bars and reported values are the mean ± SE from two replicates. (D) ATPase assays were performed in the same conditions as (A and B) and the released radioactive phosphate (Pi) was separated from non-hydrolyzed ATP by thin layer chromatography (TLC). The percentage Pi released is plotted as a function of time. ATP hydrolysis rates obtained were 0.9 μM s−1 (WT) and 0.7 μM s−1 (Δsnf5).
