Abstract
Purpose of review
This review discusses the utility of pathogen-specific antibody biomarkers for improving estimates of the population burden of waterborne infections, assessing the fraction of infections that can be prevented by specific water treatments, and understanding transmission routes and the natural history and ecology of disease in different populations (including asymptomatic infection rates).
Recent findings
We review recent literature on the application of pathogen-specific antibody response data to estimate incidence and prevalence of acute infections and their utility to assess the contributions of waterborne transmission pathways. Advantages and technical challenges associated with the use of serum versus minimally invasive salivary antibody biomarkers in cross-sectional and prospective surveys are discussed.
Summary
We highlight recent advances and challenges and outline future directions for research, development, and application of antibody-based and other immunological biomarkers of waterborne infections.
Keywords: Waterborne infections, Pathogens, Biomarkers, Antibodies, Serology, Immunoconversion, Saliva
Introduction
Waterborne infections cause an estimated two million deaths and four billion episodes of diarrheal illness per year worldwide [1]. Waterborne diseases will continue to be of broad public health importance as peri-urban populations rapidly expand at a pace that exceeds developing countries’ abilities to invest in infrastructure [2]. While most of these illnesses occur in developing countries, industrialized countries also bear a substantial burden of waterborne diseases [3]. For high-income countries, if investments in water supply and sewer systems do not enable proper maintenance and timely replacement of aging infrastructure, the risk of waterborne infections is likely to increase [4].
Waterborne disease outbreaks are defined as two or more persons experiencing a similar illness after exposure to water where epidemiologic evidence implicates water as the probable source of the outbreak [5]. Waterborne pathogens that result in human infections include bacteria (e.g., Campylobacter spp., Shigella spp.), viruses (e.g., norovirus, rotavirus), and protozoa (e.g., Cryptosporidium spp., Giardia spp.), and these pathogens may be conveyed to humans via drinking and/or recreational water transmission routes [6]. The health outcome most commonly associated with exposure to waterborne pathogens is acute gastrointestinal illness (AGI). AGI is defined in various ways, and definitions used in epidemiological research range widely [7]. One commonly used definition is as follows: diarrhea (three or more loose stools in a 24-h period), vomiting, nausea, stomach ache, fever, and/or interference with regular activities (missed time from work or school or missed regular activities as a result of illness) [8–10]. Other illnesses caused by waterborne pathogens include viral hepatitis (hepatitis A and E viruses [11]), skin and soft tissue infections and sepsis (Vibrio spp., Staphylococcus aureus [12]), primary amoebic meningoencephalitis (Naegleria fowleri [13]), and pneumonia (Legionella pneumophila [14]).
In this review, we summarize the latest evidence on use of pathogen-specific antibodies as biomarkers (defined as “any substance, structure, or process that can be measured in the body or its products and can influence or predict the incidence of outcome or disease” [15]) of infection for the waterborne pathogens that cause the greatest population burden of AGI in the USA (norovirus, Shiga toxin-producing E. coli, and Cryptosporidium spp.) [16] and in developing countries globally (rotavirus, Cryptosporidium spp., Shigella, Giardia spp., Vibrio cholerae, and Campylobacter spp.) [17, 18]. We also include hepatitis A and E viruses because these pathogens are the most common causes of feces-transmitted acute viral hepatitis worldwide (Table 1) [19, 20]. Such pathogen-specific antibody biomarkers represent promising tools to identify causative agents in population-based studies of AGI, including waterborne disease outbreak investigations, surveillance studies, and observational and randomized intervention studies to test hypotheses related to transmission routes, water treatments, and disease ecology. Because not all individuals who become infected with waterborne pathogens will experience symptoms of AGI—i.e., a waterborne infection may be asymptomatic (without clinical disease) or symptomatic (clinical disease observable) [21]—biomarkers of host immunological response can be used to identify a causative pathogenic agent and estimate symptomatic and/or asymptomatic waterborne disease burden. Knowledge of the waterborne pathogens responsible for asymptomatic infections can improve estimates of waterborne infections in source populations and advance understanding of upstream risk factors and transmission routes. Not knowing these can hinder the development of effective prevention strategies to reduce waterborne outbreaks and/or contamination events (e.g., via infrastructure improvements or other interventions prior to onset of symptoms).
Table 1.
Studies that measured specific waterborne pathogens and estimated which are responsible for the greatest population burden of waterborne infection
| Region | Data source | Top waterborne pathogens identified |
|---|---|---|
| USA | CDC Morbidity Mortality Weekly Report (MMWR) Surveillance for waterborne disease outbreaks associated with drinking water, 2011–2012 [16] | Norovirus and Shiga toxin-producing E. coli |
| CDC MMWR for Outbreaks of illness associated with recreational water, 2011–2012 [24] | Cryptosporidium spp. | |
| Developing countries | The Global Enteric Multicenter Study (GEMS)[17] | Rotavirus, Cryptosporidium spp., Shigella, Giardia spp.,a Campylobacter spp., Vibrio choleraeb |
| The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development Project (MAL-ED) [18] | Giardia spp.c | |
| Ishii et al. (2015) [19] and Hoomagle et al. (2012) [20] | Hepatitis A and E virusd |
In univariate analyses, Giardia was identified significantly more frequently in controls than in patients with moderate-to-severe diarrhea aged 12–59 months in ten of the 14 age-site strata [17]
Important in selected sites in GEMS study [17]
Giardia spp. were among the top five pathogens in terms of the highest prevalence in diarrheal and non-diarrheal stools for both the 0–11- and the 12–24-month age groups [18]
We review the challenges in measuring population burdens of infection that can be attributed to waterborne versus other transmission routes (contaminated food, hygiene, sanitation, person-to-person and animal-to-person contact). Antibodies as biomarkers of waterborne infections are then discussed to highlight their current and future utility in population-based settings. Antibody responses to specific pathogens are described as they relate to measuring immunoconversions (defined as a change from antibody negative to antibody positive in serial samples or a four-fold increase in antibody titer in serial samples), rates, and time intervals of infection. The use of antibody biomarkers in serum is presented, followed by the discussion of novel salivary antibody biomarkers and their potential to improve upon estimates of waterborne infections. The utility of antibody biomarkers for detection of acute and chronic infections in population-based settings is discussed, including how estimates of the incidence of acute short-term infections can be obtained within the context of both cross-sectional and prospective study designs. Finally, the technical challenges involved with using minimally invasive saliva samples as a matrix for the detection of pathogen-specific antibodies are presented along with future directions for salivary immunoassay work.
Challenges with Epidemiologic Estimates of Waterborne AGI in Population-Based Settings
The outcome most commonly employed in epidemiologic studies of waterborne disease is self-reported AGI symptoms. Because most AGI symptoms are self-limited, only a small proportion of the individuals who experience AGI actually seek medical care and have a stool sample submitted for testing. Furthermore, clinical diagnostic laboratories are not always able to identify a pathogenic agent responsible for AGI symptoms [22]. Thus, only a small proportion of AGI disease will be captured by studies of, or reporting systems involving, patient populations seeking a clinical diagnosis (Fig. 1). AGI symptoms are also non-specific, with numerous pathogens and transmission routes that must be investigated in order to determine the etiologic agent. These features of AGI symptoms mean that epidemiologic studies that rely upon AGI as a primary outcome may not provide an accurate estimate of the population burden of disease. The ability to determine a host's immunologic response to specific pathogens that are responsible for waterborne infections could improve the specificity and decrease the misclassification of AGI in epidemiologic studies. Biomarkers of pathogen-specific host immunologic response could improve studies of the effects of improved water treatment and/or source water protection as well as advance understanding of pathogen exposure (e.g., spatial and temporal distribution) and modifiable factors that are associated with progression from asymptomatic to symptomatic states of infection (e.g., natural history and ecology of disease) in populations. For example, objective biomarkers of asymptomatic waterborne infections have helped identify low water pressure at the faucet as an important risk factor for self-reported diarrhea in the control group of a case-control study of sporadic cryptosporidiosis [23].
Fig. 1.
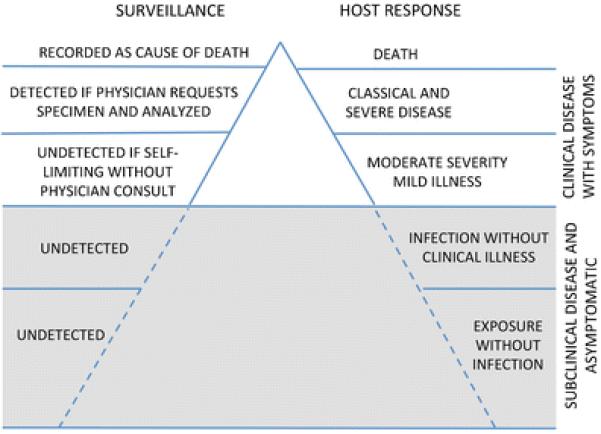
The iceberg concept of waterborne infection surveillance. Some of the “host response” information listed in Fig. 1 is adapted from Kaslow et al. [105]
Most evidence of waterborne transmission in developed countries comes from outbreaks of infectious diseases. In the USA, the Centers for Disease Control and Prevention (CDC) as well as state and local authorities investigate outbreaks and attempt to identify the source. CDC publishes the biannual Morbidity and Mortality Weekly Report on outbreaks associated with drinking and recreational water sources. For example, in 2011–2012 for drinking water, a total of 32 outbreaks were reported and associated with 431 illnesses, 102 hospitalizations, and 14 deaths [16]. For recreational water in 2011–2012, there were 90 outbreaks that resulted in at least 1788 cases, 95 hospitalizations, and one death [24].
Knowledge of the pathogen-specific etiology of waterborne infections would help identify different risk factors and transmission routes, which can improve the evidence base for decision making about management and prevention strategies. A classic example of this is the massive waterborne outbreak of cryptosporidiosis in Milwaukee in 1993 when the chlorine-based disinfectant used had little effect on Cryptosporidium parvum oocysts and the drinking water treatment plants consequently had to investigate alternative disinfectants such as UV light [25]. Another example is a study of the presence of enteric viruses in non-disinfected drinking water from municipal wells and their relation with community incidence of AGI [26]. In this study, the authors noted a positive association between norovirus genogroup I (GI) and AGI. But, the associations between the presence of other enteric viruses—adenovirus and echovirus serotypes—and AGI were not statistically significant. This lack of association could be due to misclassification and/or the non-specificity of AGI as an outcome in epidemiologic studies (e.g., potential influence of measurement error due to participant self-reporting of AGI symptoms).
Waterborne outbreaks usually occur from causative factors such as weather events, wastes from animals, agriculture, or humans, and failures in water treatment [27]. Drinking water-associated outbreaks are often caused by contaminated source waters, inadequacies in treatment, or contamination occurring within the distribution system [28]. Whereas, recreational water-associated outbreaks have been attributed to swimming in waters impacted by inadequate chlorination or other disinfection (swimming pools) [29], fecal contamination shed by swimmers (swimming pools and natural waters) [30], runoff from publicly owned treatment work (POTW) wastewater effluents, sanitary and combined sewer overflows of untreated sewage, private on-site septic systems, agricultural production, and wildlife [31].
Most cases of waterborne infections are sporadic or diffuse, low-level outbreaks. Ingestion of waterborne pathogens can also result in a completely asymptomatic infection depending on the interplay of pathogen-specific and host-specific factors, such as a pathogen's virulence and a host's immune response [32]. They may be caused by deficiencies in drinking water treatment, resulting in contamination with waterborne pathogens, and transmission to consumers [33]. Waterborne pathogens that are resistant to chlorination (especially Cryptosporidium spp.) [34] or physical removal (especially viruses) can pass through the water treatment barrier and contaminate tap water even when water quality indicators based on surrogate bacteria (total and/or fecal coliforms, E. coli) are within the regulatory limits [35]. Viruses, such as noroviruses, can filter through the soil, contaminate shallow groundwater sources and present a health risk in drinking water systems that are groundwater supplied and do not use chemical disinfection [26]. Individual sporadic cases of AGI usually cannot be linked to a specific source in the framework of routine surveillance, contributing to the underestimation of waterborne infections in the population.
Antibody Biomarkers of Waterborne Infection
Specific antibody responses can be used as biomarkers of infection in epidemiological studies to estimate the prevalence and incidence of infections and to assess the contribution of waterborne transmission. Different pathogens result in different temporal distributions of antibody response and infection. Both symptomatic and asymptomatic infections typically cause an antibody response in the host [33]. A preexisting antibody response can be a factor affecting host's susceptibility to re-infection or the probability of developing symptoms if infection occurs [36]. The presence of antibodies specific to the pathogen of interest in biological samples (e.g., serum, saliva, stool, breast milk) is an indication of current or prior infection [33]. The major immunoglobulin isotypes (IgG, IgA, IgM) have different utility as estimates of population disease frequency and burden. Single time-point measurements of pathogen-specific IgG have utility as an estimate of historical/prior exposure or prevalent infection, whereas IgA and/or IgM has utility as an estimate of acute phase or incident infection [37, 38]. Immunoconversion is used to detect incident infections in prospective survey settings. This change from an antibody-negative sample to an antibody-positive sample in a time series of two or more samples, or a fourfold increase in antibody titer in a time series of two or more antibody-positive samples, is used to measure new, acute cases in a defined population over a defined time period [39–41].
Serologic Antibody Response
Serum is the most accurate and widely used matrix to monitor population immune responses to pathogens. Sera can be collected by sampling populations, or residual blood banks can be used. However, there are significant drawbacks to both since blood collection requires trained individuals to visit participants [42] and may be cost prohibitive along with low response rates that have been shown in Europe due to the invasive nature of blood collection [43, 44]. Its application in prospective studies and especially in studies involving children is problematic due to high attrition and low compliance [45]. Relying on previously collected samples from sera banks overcomes these issues; however, they are usually anonymous with limited data available on the patient and, importantly, their background as it pertains to water, sanitation, and hygiene-related behaviors and activities [46]. However, a number of studies have successfully used seroepidemiological methods in the context of waterborne disease [47, 48]. Frost et al. found that people who live in cities using surface-derived drinking waters had an increased risk of Cryptosporidium infection compared to those using drinking water from municipal groundwater sources [47]. And, in the context where sanitation conditions are poor and clean water supplies are limited, Priest et al. found IgG antibody responses during Cryptosporidium infections with C. parvum, Cryptosporidium felis, and Cryptosporidium meleagridis and with four different subtypes of Cryptosporidium hominis [48].
Salivary Antibody Response
The utility of novel salivary antibody biomarkers as a measure of host immune response to specific pathogens has the potential to improve upon estimates of waterborne infections that rely on invasive collection of serum. Saliva collection is minimally invasive and can be self-collected and returned by mail [49••], allowing for a larger sampling of the population than is possible with serum. Saliva is a mixture of secretions from salivary glands. Oral fluid contains saliva (enriched with secre-tory IgA) and crevicular fluid (flows from between the gum margins and teeth) and is enriched with serum antibodies [50]. Some oral fluid sampling techniques are specifically designed to collect samples enriched with crevicular fluid for measurements of systemic antibody responses [51, 52, 53•].
Salivary assays have been used to identify various viral, bacterial, and parasitic infections [54] (see Table 2). Measuring antibodies in saliva is appropriate for both children and adults and is suitable for population-based surveillance settings [40]. Salivary immunoassays have been developed for pathogens such as Helicobacter pylori, Toxoplasma gondii, Cryptosporidium, and noroviruses [52••]. Griffin et al. (2011) applied the Luminex xMAP microsphere-based technology (Luminex Corp., Austin, TX) assay to measure antibodies to multiple pathogens within a single saliva sample volume [52••]. The Norwalk virus assay developed in Griffin et al. (2011) was subsequently validated using samples from a human volunteer challenge study [53•]. A similar salivary immunoassay is being applied to measure the incidence of norovirus infections following recreational water exposures at beaches in Puerto Rico, Iowa, and Wisconsin where saliva has been collected as part of the Environmental Protection Agency's National Epidemiologic and Environmental Assessment of Recreational Water Study [55].
Table 2.
Immunological biomarkers of infection for waterborne pathogens that are responsible for the greatest global burden of acute gastrointestinal illness (AGI)
| Pathogen of interest | Specimen | Immunologic biomarker response | Reference |
|---|---|---|---|
| Cryptosporidium spp. | Serum | IgG antibody | Priest, J. W., et al. [106]; Chappell, C. L., et al. [60]; Crump, J. A., et al. [107]; Sarkar, R., et al. [108]; Becker, D. J., et al. [109]; Checkley, W., et al. [110] |
| Saliva | IgG and IgA antibody | Cozon, G., et al. [111]; Moss, D. M., et al. [69]; Egorov, A. I., et al. [112]; Griffin, S. M., et al. [52••]; | |
| Campylobacter | Serum | IgG, IgM, and IgA antibodies | Ang, C. W., et al.; [93]; Teunis, P. F., et al. [81]; Rokosz-Chudziak, N. and W. Rastawicki [113]. |
| Stool | Cytokines (IL-1β, IL-6, IL-8, TNF-α, and IFN-γ), IgA antibodies | Tribble, D. R., et al. [114]; Islam, D., et al. [115]; | |
| Saliva | IgG and IgA antibodies (responses to acid-glycine extracts of C. jejuni strain 81116 and an aflagellate mutant, and a whole-cell R2 sonicate) | Cawthraw, S. A., et al. [116] | |
| Giardia intestinalis | Serum | IgG and IgA antibodies | Crump, J. A., et al. [107]; Jiménez, J. C., et al. [117]; Priest, J. W., et al. [66]; Moss, D. M., et al. [68] |
| Saliva | sIgA, IgA, and IgG antibody (responses against G. duodenalis) | Rodriguez, O. L., et al. [118]; El-Gebaly, N. S., et al. [119] | |
| Hepatitis A virus | Serum | IgM and IgG antibodies | Vitral, C. L., et al. [11]; Hundekar, S., et al. [120] |
| Saliva | IgM and IgG antibodies | Laufer, D. S., et al. [121]; Ochnio, J. J., et al. [122]; Morris-Cunnington, M. C., et al. [49••]; Tourinho, R. S., et al. [123] | |
| Hepatitis E virus | Serum | IgG and IgM antibody, cytokines (IL-5, IL-6, IL-8, IL-10, IL-2, IFN-γ,TNF-α,TGF-β1, IL-1β | Adjei, A. A., et al. [124]; Pas, S. D., et al. [125]; Wu, W. C., et al. [38]; Kumar, A., et al. [126]; Gu, G., et al. [127]; Cong, W., et al. [37]; Heaney, C. D., et al. [128], Kmush, B. L., et al. [129] |
| Norovirus | Serum | IgG and IgA antibodies, cytokines (IL-1, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, and IL-12, IFN-β,TNF-α) | Erdman, D. D., et al. [64]; Monroe, S. S., et al. [39]; Moe, C. L., et al. [40]; Lindesmith, L., et al. [58]; Crump, J. A., et al. [107]; Newman, K. L., et al. [95] |
| Stool | IgA antibody | Iritani, N., et al. [130]; Ramani, S., et al. [131] | |
| Saliva | IgA and IgG antibodies | Moe, C. L., et al. [40]; Lindesmith, L., et al. [59]; Lindesmith, L., et al. [58]; Griffin, S. M., et al. [52••]; Griffin, S. M., et al. [53•] | |
| Rotavirus | Serum | IgM, IgA, and IgG antibodies, cytokines (IFN-γ, TNF-α, IL-8, and IL-10) | Grimwood, K., et al. [132]; Azim, T., et al. [133]; Xu, J., et al. [134]; Premkumar, P., et al. [135]; Sindhu, K. N., et al. [136]; Moon, S. S., et al. [137] |
| Stool | IgM, IgA, and IgG antibodies | Stals, F., et al. [138]; Grimwood, K., et al. [132]; Azim, T., et al. [133] | |
| Saliva | IgM, IgA, and IgG antibodies | Stals, F., et al. [138]; Grimwood, K., et al. [132]; Aiyar, J., et al. [139]; Friedman, M. G., et al. [140]; |
|
| Shiga toxin-producing | Serum | IgG antibodies against 51 O serogroup strains, B subunit of Stx2 and Stx1 | Ludwig, K., et al. [141]; Kulkarni, H., et al. [142]; Fernández-Brando, R. J., et al. [143]; Guirro, M., et al. [144] |
| Escherichia coli | Saliva | IgM and IgA antibodies | Ludwig, K., et al. [145]; Chart, H., et al. [146] |
| Shigella | Serum | IgA, IgM, and IgG subtypes to S. sonnei O-antigen, IgA and IgG antibodies to S. flexneri 2a lipopolysaccharide, total IgA antibody-secreting cells (ASC), and anti-LPS IgA ASC, cytokines (IFN-γ,TNF-α,TNF-β, IL-4, IL-6, TGF-β) | Van De Verg, L. L., et al. [147]; Raqib, R., et al. [148]; Rasolofo-Razanamparany, V., et al. [149]; Levine, M. M., et al. [150]; Muhsen, K., et al. [151]; Thompson, C. N., et al. [152] |
| Stool | Cytokines (TNF-α, IL-6) | Azim, T., et al. [153] | |
| Saliva | IgA antibody | Schultsz, C., et al. [154]; | |
| Vibrio cholerae | Serum | IgA and IgG antibodies, IgG, IgM, and IgA ASC | Chowdhury, F., et al. [155]; Johnson, R. A., et al. [156]; Fujii, Y., et al. [157]; Khan, A. I., et al. [158] |
| Stool | IgA antibody | Qadri, F., et al. [159] | |
| Saliva | IgA antibody | Jertborn, M., et al. [160] |
An important challenge in using saliva to measure immunologic responses is the greater inter- and intra-individual variability in saliva composition and immunoglobulin levels. While saliva contains a high level of secretory IgA (SIgA) antibodies, there can be significant diurnal, age, and oral health-related variability [56], making these factors important to consider in community-based field studies. The salivary concentrations of IgG and IgM isotypes are lower than in serum. Thus, a salivary antibody assay targeting IgG has to be sensitive enough to quantify low-intensity antibody responses. Typically, it is necessary to assay saliva at relatively low dilutions, where matrix effects (e.g., inhibition, high background signal) can be pronounced in some pathogen-specific antibody assays [57]. For each pathogen-specific antibody target, it is critical to optimize the conditions that may influence assay performance and sensitivity and specificity [53•].
There is scant evidence on the temporal patterns of salivary antibody responses to infection with a specific pathogen (peak levels and rates of decline for different antibody isotypes). Our current understanding of generalized trajectories (Fig. 2) comes from prospective studies using serum or saliva from individuals with confirmed infections, such as volunteer challenge studies for norovirus [40, 58, 59], Cryptosporidium [60], Giardia lamblia [61, 62], and Shigella [63]. The pattern of antibody isotypes may be used in diagnostic and research settings to provide information on the infection state (acute versus convalescent) and to assess the timing of infection [33]. Typically, the IgA and/or IgM response to a waterborne pathogen ramps up before the IgG response [36, 58, 59]. The generalized trajectories of different antibody isotype levels during a transient acute infection from a waterborne pathogen are depicted in Fig. 2. After the convalescent stage, IgG pathogen-specific antibodies may remain detectable for weeks to years, depending on the causative agent, and may remain elevated above preinfection levels [36, 64]. There can be vast differences in these temporal patterns of antibody responses depending on the pathogen causing the infection. Thus, an area of future work is to develop population-based antibody infection curves for specific waterborne pathogens.
Fig. 2.
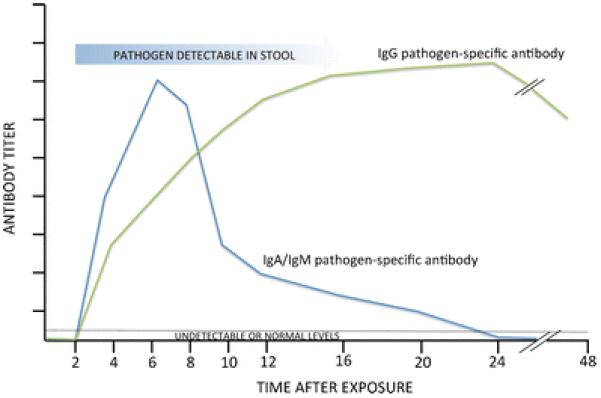
Temporal pattern of antibody responses during infection with a waterborne pathogen
Platforms and Assay Types
Various immunoassay platforms have different costs, quantitation levels, dynamic ranges, and multiplexing potentials [65]. The most basic of these platforms is the indirect enzyme immunoassay; however, the low through-put and high sample volume requirements make it less desirable for population-based analyses where multiple pathogens are being analyzed and sample volume is limited. Multiplex immunoassays, such as those based on the Luminex (Luminex Corp., Austin, TX) microbead suspension fluorescence immunoassay platform, require a low sample volume to analyze multiple pathogen-specific antibody analytes simultaneously. They are also less labor intensive because more data are generated per test/analyte and thus are more cost-effective [52••, 53•, 66–69]. Another immunoassay platform that is used and allows multiplexing is the Meso Scale Discovery (MSD; Rockville, M D) electrochemiluminescence (ECL) platform. Platforms that facilitate multiplexing can be used to expand the range of available options for testing the signal of pathogen-specific antibody responses as well as background signals. The adjustment of the pathogen-specific antibody signal for background signals, such as those produced by total IgA or total IgG or by antigen tags such as glutathione-S-transferase (used during antigen purification), can improve the performance of antibody assays [52••, 53•]. Multiplexing of these target signals can also reduce excess use of biospecimen sample volume because all signals can be measured in one sample volume in a single reaction well. Thus, multiplexing testing platforms can facilitate a broader application of antibody testing of serum and/or saliva biospecimens in population-based epidemiologic investigations of diverse waterborne pathogens.
Applications of Pathogen-Specific Antibody Biomarkers in Population-Based Studies of Waterborne Infections
To improve current epidemiologic estimates of AGI from waterborne pathogens in population-based settings, pathogen-specific antibody biomarkers can be used. For chronic infections, antibody responses can be positive or negative and can be validated against diagnostic tests. The proportion of IgG-positive results in serum or saliva can serve as a direct measure of infection prevalence in the population [70, 71]. In contrast, for acute short-term infections, such as noroviruses and Cryptosporidium, the presence of pathogen-specific antibodies in serum or saliva may indicate an ongoing infection or more commonly a past infection with or without symptoms. Thus, the concept of “positive” antibody response to an acute short-term infection or seroprevalence of positive responses often reflects the proportion of results above an arbitrary threshold, such as a detection limit of the method or by standardizing response intensities to the response of a reference sample of positive control sera [72–75] or saliva.
One approach to estimating incidence of acute infections using antibody data is to use immunoconversion in prospective study settings as a marker of new infections. The sensitivity and specificity of an immunoconversion test are related to its ability to detect infections that occur during the interval between two sampling dates. In prospective studies, biological sampling (serum or saliva) can be combined with symptom diaries to produce information on the association of certain infections with specific types of symptoms and/or the association of exposures with infections or interventions (designed to reduce exposure) with a lack of symptoms [76].
Prior studies have used pathogen-specific antibody markers and demonstrated their ability to identify waterborne infections that were more widespread than previously appreciated. In the massive Cryptosporidium outbreak in Milwaukee in April 1993, a retrospective analysis was conducted with banked serum specimens from children that had routine lead level surveillance in blood from March to May of that year and showed a seroprevalence increase from 15–17 % to 82–87 % for levels of IgG antibody against the immunodominant Triton-17 and 27 kDa C. parvum antigens [77]. This demonstrated that the outbreak had affected a greater proportion of the population with infection when accounting for both symptomatic and asymptomatic infections than the previous estimate of 26 % that only surveyed the population using the cryptosporidiosis case definition (watery diarrhea) [78]. Teunis et al. applied these approaches in the European Union to estimate seroconversion rates for Campylobacter infections and found that they were several orders of magnitude higher than the notification rates, reflecting both detection deficits in the surveillance and the reality that these enteric infections often remain asymptomatic [79]. Frost et al. used serum antibodies to Cryptosporidium from a population in Hungary to determine that those using groundwater had significantly lower serological responses than those using conventionally filtered and disinfected surface water and found that riverbank filtration may be an effective alternative treatment to reduce Cryptosporidium exposures and infections for individuals using surface water sources [80]. Tollestrup et al. focused on non-outbreak settings where a low probability of outbreak detection should be expected and found a significant association for residents in the River Valley of New Mexico using onsite wastewater systems combined with private wells to have a strong response to the 27 kDa Cryptosporidium antigen [75]. And lastly, in the first postal population-based survey that used saliva, Morris-Cunnington et al. used approximately 5500 self-collected oral fluid samples along with a questionnaire of demographic and social information to successfully demonstrate that antibody prevalence data along with risk factor data can be used to assess the population-based immunity to common viral infections in England and Wales [49••].
Such applications of immunological biomarkers in epidemiologic studies also can improve knowledge of the temporal patterns of antibody responses, which can be used to extrapolate incidence estimates based on cross-sectional data on pathogen-specific antibody responses in the population [79, 81, 82]. Others have expanded this approach using parametric statistical models [67, 83–85] to determine incidence of infection based on pathogen-specific antibody results from a single cross-sectional sampling time. The person-to-person variability in antibody responses to a specific pathogen and limited data on temporal patterns of antibody responses in various populations affect the precision of such estimates. A pattern of antibody responses may also be affected by the number of prior infections and the time interval since the previous infection. This may further limit the applicability of the available antibody pattern data to populations with comparable epidemiological characteristics or to research questions focused on intra-individual variability in antibody responses over time.
In low-income communities where there is less developed drinking water and wastewater infrastructure and individuals may experience repeated exposures to multiple waterborne pathogens, the application of immunological biomarkers can be used as a monitoring and evaluation tool for infrastructure and point-of-use interventions. The multiplex immunoassay methodology targeting salivary IgG and IgA responses to potentially waterborne pathogens [52••] can be applied as a minimally invasive and objective exposure and outcome screening tool to assess the efficacy of interventions designed to reduce pathogen exposure and/or AGI illness within a specified population. Such multiplex pathogen antibody measurements could improve the evaluation and prioritization of a range of water, sanitation, hygiene, and health programs and interventions. Integration of these biomarkers into monitoring activities for the Sustainable Development Goals recently adopted at the 2015 UN Summit (https://sustainabledevelopment.un.org/topics) could improve the evidence base for improved health outcomes related to Goal 6, which is to “by 2030, achieve access to adequate and equitable sanitation and hygiene for all and end open defecation” (Target 6.2) [86].
Biomarkers of pathogen-specific antibody response can also be used to improve monitoring and evaluation of vaccination coverage for specific waterborne infections. Several waterborne pathogen vaccines for which antibody response data can be generated include Shigella [87], rotavirus [88], cholera [89, 90], and hepatitis A [91] and E [92], among others. Such pathogen-specific antibody response biomarkers have particular utility in remote, resource-limited population-based settings because they can provide objective measures of vaccination coverage when paper-based records and/or recall of vaccination history is lacking.
Challenges and Perspectives for Future Work
Pathogen-specific antibody assays represent a promising tool for understanding the relative contribution of waterborne versus other pathways to infectious disease burden in population-based settings. However, assays based on invasive serum specimens may fail to capture a majority of cases in population-based field studies. Because saliva swabs can be self-administered and returned by mail [49••], salivary antibody assays may increase participation in surveys of potentially waterborne infections in populations that are difficult to reach, including children, pregnant women, and individuals living in remote, resource-limited settings. This may facilitate a more fine-scale, spatiotemporal study of the ecology and natural history of waterborne disease, including elucidation of optimal points of intervention to prevent waterborne pathogen transmission.
While such minimally invasive pathogen-specific salivary antibody biomarkers are promising, challenges remain in their broad application to diverse pathogen exposures and infections. Not all pathogens elicit a robust systemic or salivary antibody response. Additionally, a majority of waterborne infections may be asymptomatic and not result in adverse health effects. Therefore, the incidence of infections estimated from cross-sectional antibody data may not be representative of disease burden but only reflect recent or historical exposure to a pathogen [93]. Nevertheless, cross-sectional antibody response data can provide an improved estimate of human exposure to specific pathogens and can be used as an epidemiological tool to estimate the contribution of waterborne versus other pathways to the total infection pressure. However, the underlying infection and immune response to the pathogen must be considered in the interpretation of cross-sectional seroprevalence estimates and depend on whether the infection results in lifetime immunity following one exposure or the infection is acute and immunity wanes following exposure.
The detection of cytokines in serum and saliva also presents an opportunity to measure the onset of waterborne infections. However, cytokines are not capable of identifying a specific causative agent; rather, they are more generic biomarkers of infection. The hallmark for a viral infection begins with a wave of cytokine production [94], and their presence can be employed as a marker of infection (Table 2). Cytokine levels in serum of individuals infected with norovirus that were shown to be significantly increased included IFN-gamma, interleukin 6 (IL-6), IL-8, IL-12p70, MCP-1, and TNF-alpha 2 days following exposure [95]. Evidence has shown that the elevation of cytokines in a newborn's salivary gland epithelium promotes secretory immunity [96]. Proinflammatory cytokines can upregulate the polymeric Ig receptor (pIgR), including IL-17, which is particularly abundant at mucosal sites [97]. The extracellular part of pIgR is essential for resistance against proteolytic degradation of the secretory component of IgA (SIgA) found in saliva and the gut mucosa [98]. A challenge in using cytokines in saliva is to determine if there is a serum-saliva association, for which there is currently limited evidence [99]. The most likely hypothesis is that much of the variation in salivary cytokines (e.g., IL-1b, TNF-α, IL-6, IL-8) may be due to inflammatory processes in the mouth caused by poor oral health [100] and/or other disease processes [101–103]. However, there could be specific hyper-inflammatory physiological states (systemic infection/sepsis, burns, etc.) when more of the variance in salivary levels of cytokines could be due to systemic circulating cytokine levels [99]. IL-6, which has a major role in the regulation of inflammatory processes, was found to be elevated in concentration in both the saliva and serum of inflammatory bowel disease patients when compared to reference persons [104]. An area for future study is identifying if a specific waterborne pathogen generates a unique or predictive cytokine profile that is observable in both saliva and serum.
Conclusion
The ability to estimate waterborne infections via measurements of host immunological response at the population level is improving as technological and analytical advancements are made. Diagnostic advancements are enabling a paradigm shift in how waterborne infections can be measured, not just in clinical settings or outbreak settings but also more widely as tools for population-based screening of incidence and prevalence. The measurement of salivary antibody responses to specific pathogens as biomarkers of waterborne infection holds great potential to expand surveillance to reach larger numbers of people in diverse population-based settings. Future work lies in the development of sensitive and specific multiplexed serum and salivary immunoassays to measure exposures to, and infections with, specific waterborne pathogens.
Footnotes
Compliance with Ethical Standards
Conflict of Interest Douglas A. Granger is the founder and chief scientific and strategy advisor at Salimetrics LLC and SalivaBio LLC, and these relationships are managed by the policies of the committees on conflict of interest at the Johns Hopkins University School of Medicine and the University of California at Irvine. All other authors declare no conflict of interest.
Human and Animal Rights and Informed Consent This is a review article which does not report new results of human or animal subjects performed by the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Organization WH. UN-Water global annual assessment of sanitation and drinking-water (GLAAS) 2012 report: the challenge of extending and sustaining services. 2012.
- 2.Ali SI. Alternatives for safe water provision in urban and peri-urban slums. J Water Health. 2010;8(4):720–34. doi: 10.2166/wh.2010.141. [DOI] [PubMed] [Google Scholar]
- 3.Shortridge JE, Guikema SD. Public health and pipe breaks in water distribution systems: analysis with internet search volume as a proxy. Water Res. 2014;53:26–34. doi: 10.1016/j.watres.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Gargano JW, et al. Acute gastrointestinal illness following a prolonged community-wide water emergency. Epidemiol Infect. 2015;143(13):2766–76. doi: 10.1017/S0950268814003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn BG, et al. Surveillance for waterborne-disease outbreaks associated with drinking water—United States, 2001-2002. Morb Mortal Wkly Rep Surveill Summ. 2004;53(SS-8):23–45. [PubMed] [Google Scholar]
- 6.World Health Organization . Guidelines for drinking-water quality: recommendations. Vol. 1. World Health Organization; Geneva: 2004. [Google Scholar]
- 7.Roy SL, Scallan E, Beach MJ. The rate of acute gastrointestinal illness in developed countries. J Water Health. 2006;4(Suppl 2):31–69. doi: 10.2166/wh.2006.017. [DOI] [PubMed] [Google Scholar]
- 8.Colford JM, Jr, et al. The Sonoma water evaluation trial: a randomized drinking water intervention trial to reduce gastrointestinal illness in older adults. Am J Public Health. 2009;99(11):1988–95. doi: 10.2105/AJPH.2008.153619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFelice NB, Johnston JE, Gibson JM. Acute gastrointestinal illness risks in North Carolina community water systems: a methodological comparison. Environ Sci Technol. 2015;49(16):10019–27. doi: 10.1021/acs.est.5b01898. [DOI] [PubMed] [Google Scholar]
- 10.Wymer LJ, Wade TJ, Dufour AP. Equivalency of risk for a modified health endpoint: a case from recreational water epidemiology studies. BMC Public Health. 2013;13:459. doi: 10.1186/1471-2458-13-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitral CL, et al. Hepatitis A and E seroprevalence and associated risk factors: a community-based cross-sectional survey in rural Amazonia. BMC Infect Dis. 2014;14:458. doi: 10.1186/1471-2334-14-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esiobu N, et al. High numbers of Staphylococcus aureus at three bathing beaches in South Florida. Int J Environ Health Res. 2013;23(1):46–57. doi: 10.1080/09603123.2012.699027. [DOI] [PubMed] [Google Scholar]
- 13.Ashbolt NJ. Microbial contamination of drinking water and human health from community water systems. Curr Environ Health Rep. 2015;2(1):95–106. doi: 10.1007/s40572-014-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkinham JO, 3rd, et al. Epidemiology and ecology of opportunistic premise plumbing pathogens: Legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa. Environ Health Perspect. 2015;123(8):749–58. doi: 10.1289/ehp.1408692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization . Environmental Health Criteria. World Health Organization; Geneva: 2001. Biomarkers in risk assessment: Validity and validation. [Google Scholar]
- 16.Beer KD, et al. Surveillance for waterborne disease outbreaks associated with drinking water—United States, 2011-2012. MMWR Morb Mortal Wkly Rep. 2015;64(31):842–8. doi: 10.15585/mmwr.mm6431a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–22. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 18.Platts-Mills JA, et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health. 2015;3(9):e564–75. doi: 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii K, et al. Epidemiological and genetic analysis of a 2014 outbreak of hepatitis A in Japan. Vaccine. 2015;33(45):6029–36. doi: 10.1016/j.vaccine.2015.04.061. [DOI] [PubMed] [Google Scholar]
- 20.Hoofnagle JH, Nelson KE, Purcell RH. Hepatitis E. N Engl J Med. 2012;367(13):1237–44. doi: 10.1056/NEJMra1204512. [DOI] [PubMed] [Google Scholar]
- 21.Leclerc H, Schwartzbrod L, Dei-Cas E. Microbial agents associated with waterborne diseases. Crit Rev Microbiol. 2002;28(4):371–409. doi: 10.1080/1040-840291046768. [DOI] [PubMed] [Google Scholar]
- 22.Kubota K, et al. The human health burden of foodborne infections caused by Campylobacter, Salmonella, and Vibrio parahaemolyticus in Miyagi Prefecture. Jpn Foodborne Pathog Dis. 2008;5(5):641–8. doi: 10.1089/fpd.2008.0092. [DOI] [PubMed] [Google Scholar]
- 23.Hunter PR, et al. Self-reported diarrhea in a control group: a strong association with reporting of low-pressure events in tap water. Clin Infect Dis. 2005;40(4):e32–4. doi: 10.1086/427750. [DOI] [PubMed] [Google Scholar]
- 24.Hlavsa MC, et al. Outbreaks of illness associated with recreational water—United States, 2011-2012. MMWR Morb Mortal Wkly Rep. 2015;64(24):668–72. [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson AM, et al. UVinactivation of Cryptosporidium hominis as measured in cell culture. Appl Environ Microbiol. 2005;71(5):2800–2. doi: 10.1128/AEM.71.5.2800-2802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borchardt MA, et al. Viruses in nondisinfected drinking water from municipal wells and community incidence of acute gastrointestinal illness. Environ Health Perspect. 2012;120(9):1272–9. doi: 10.1289/ehp.1104499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster CJ, et al. Infectious disease outbreaks related to drinking water in Canada, 1974-2001. Can J Public Health. 2005;96(4):254–8. doi: 10.1007/BF03405157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risebro HL, et al. Fault tree analysis of the causes of waterborne outbreaks. J Water Health. 2007;5(Suppl 1):1–18. doi: 10.2166/wh.2007.136. [DOI] [PubMed] [Google Scholar]
- 29.Stafford R, et al. A community outbreak of Cryptosporidium infection associated with a swimming pool complex. Commun Dis Intell. 2000;24(8):236–9. doi: 10.33321/cdi.2000.24.38. [DOI] [PubMed] [Google Scholar]
- 30.Cope JR, et al. Preventing community-wide transmission of Cryptosporidium: a proactive public health response to a swimming pool-associated outbreak—Auglaize County, Ohio. USA Epidemiol Infect. 2015;143(16):3459–67. doi: 10.1017/S0950268815000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoen ME, Soller JA, Ashbolt NJ. Evaluating the importance of faecal sources in human-impacted waters. Water Res. 2011;45(8):2670–80. doi: 10.1016/j.watres.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 32.Damania B, Dittmer DP. What lies within: coinfections and immunity. Cell Host Microbe. 2014;16(2):145–7. doi: 10.1016/j.chom.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casemore D. Towards a US national estimate of the risk of endemic waterborne disease—sero-epidemiologic studies. J Water Health. 2006;4(Suppl 2):121–63. doi: 10.2166/wh.2006.021. [DOI] [PubMed] [Google Scholar]
- 34.Chalmers RM, Giles M. Zoonotic cryptosporidiosis in the UK—challenges for control. J Appl Microbiol. 2010;109(5):1487–97. doi: 10.1111/j.1365-2672.2010.04764.x. [DOI] [PubMed] [Google Scholar]
- 35.Locas A, et al. Virus occurrence in municipal groundwater sources in Quebec Canada. Can J Microbiol. 2007;53(6):688–94. doi: 10.1139/W07-034. [DOI] [PubMed] [Google Scholar]
- 36.Krain LJ, Nelson KE, Labrique AB. Host immune status and response to hepatitis E virus infection. Clin Microbiol Rev. 2014;27(1):139–65. doi: 10.1128/CMR.00062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cong W, et al. Seroprevalence of hepatitis E virus among pregnant women and control subjects in China. J Med Virol. 2015;87(3):446–50. doi: 10.1002/jmv.24058. [DOI] [PubMed] [Google Scholar]
- 38.Wu WC, et al. Application of serologic assays for diagnosing acute hepatitis E in national surveillance of a nonendemic area. J Med Virol. 2014;86(4):720–8. doi: 10.1002/jmv.23785. [DOI] [PubMed] [Google Scholar]
- 39.Monroe SS, et al. Detection of antibody to recombinant Norwalk virus antigen in specimens from outbreaks of gastroenteritis. J Clin Microbiol. 1993;31(11):2866–72. doi: 10.1128/jcm.31.11.2866-2872.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moe CL, et al. Diagnosis of norwalk virus infection by indirect enzyme immunoassay detection of salivary antibodies to recombinant norwalk virus antigen. Clin Diagn Lab Immunol. 2004;11(6):1028–34. doi: 10.1128/CDLI.11.6.1028-1034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leon J, et al. In: Immunology of norovirus infection, in immunity against mucosal pathogens. Vajdy M, editor. Springer; Netherlands: 2008. pp. 219–62. [Google Scholar]
- 42.Emmons W. Accuracy of oral specimen testing for human immunodeficiency virus. Am J Med. 1997;102(4 A):15–20. doi: 10.1016/s0002-9343(97)00033-8. [DOI] [PubMed] [Google Scholar]
- 43.De Melker HE, et al. Non-participation in a population-based seroprevalence study of vaccine-preventable diseases. Epidemiol Infect. 2000;124(2):255–62. doi: 10.1017/s0950268899003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sethi D, et al. A study of infectious intestinal disease in England: plan and methods of data collection. Commun Dis Public Health. 1999;2(2):101–7. [PubMed] [Google Scholar]
- 45.McMurtry CM, et al. Far from “just a poke”: common painful needle procedures and the development of needle fear. Clin J Pain. 2015;31(10 Suppl):S3–S11. doi: 10.1097/AJP.0000000000000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osborne K, et al. Ten years of serological surveillance in England and Wales: methods, results, implications and action. Int J Epidemiol. 2000;29(2):362–8. doi: 10.1093/ije/29.2.362. [DOI] [PubMed] [Google Scholar]
- 47.Frost FJ, et al. Serological responses to Cryptosporidium antigens among users of surface- vs ground water sources. Epidemiol Infect. 2003;131(3):1131–1138. doi: 10.1017/s0950268803001341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Priest JW, et al. Longitudinal analysis of cryptosporidium species-specific immunoglobulin G antibody responses in Peruvian children. Clin Vaccine Immunol. 2006;13(1):123–31. doi: 10.1128/CVI.13.1.123-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Morris-Cunnington MC, et al. A population-based seroprevalence study of hepatitis a virus using oral fluid in England and Wales. Am J Epidemiol. 2004;159(8):786–94. doi: 10.1093/aje/kwh107. [One of the first population-based surveillance studies involving self-collection and postal return of oral fluids samples.] [DOI] [PubMed] [Google Scholar]
- 50.Brandtzaeg P. Do salivary antibodies reliably reflect both mucosal and systemic immunity? Ann N Y Acad Sci. 2007;1098:288–311. doi: 10.1196/annals.1384.012. [DOI] [PubMed] [Google Scholar]
- 51.McKie A, Vyse A, Maple C. Novel methods for the detection of microbial antibodies in oral fluid. Lancet Infect Dis. 2002;2(1):18–24. doi: 10.1016/s1473-3099(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 52••.Griffin SM, et al. Development of a multiplex microsphere immunoassay for the quantitation of salivary antibody responses to selected waterborne pathogens. J Immunol Methods. 2011;364(1–2):83–93. doi: 10.1016/j.jim.2010.11.005. [This study provides the first stage of a pilot, proof-of-concept project to develop a non-invasive salivary antibody technique for surveillance of waterborne infections.] [DOI] [PubMed] [Google Scholar]
- 53•.Griffin SM, et al. Application of salivary antibody immunoassays for the detection of incident infections with Norwalk virus in a group of volunteers. J Immunol Methods. 2015;424:53–63. doi: 10.1016/j.jim.2015.05.001. [This study demonstrated that the use of salivary antibodies in conjunction with recombinant Norwalk virus P particles may enable inexpensive and non-invasive surveillance of incident Norwalk virus infections in prospective epidemiological studies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaka-ur-Rab Z, et al. Evaluation of salivary anti-Salmonella typhi lipopolysaccharide IgA ELISA for serodiagnosis of typhoid fever in children. Arch Dis Child. 2012;97(3):236–8. doi: 10.1136/adc.2011.300622. [DOI] [PubMed] [Google Scholar]
- 55.Augustine SA, et al. Development and application of a salivary antibody 6-plex immunoassay to determine human exposure to environmental pathogens. American Chemical Society 250th National Meeting and Exposition; Boston, MA. 2015. [Google Scholar]
- 56.Dimitriou L, Sharp NC, Doherty M. Circadian effects on the acute responses of salivary cortisol and IgA in well trained swimmers. Br J Sports Med. 2002;36(4):260–4. doi: 10.1136/bjsm.36.4.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandin A, et al. High salivary secretory IgA antibody levels are associated with less late-onset wheezing in IgE-sensitized infants. Pediatr Allergy Immunol. 2011;22(5):477–81. doi: 10.1111/j.1399-3038.2010.01106.x. [DOI] [PubMed] [Google Scholar]
- 58.Lindesmith L, et al. Cellular and humoral immunity following Snow Mountain virus challenge. J Virol. 2005;79(5):2900–9. doi: 10.1128/JVI.79.5.2900-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9(5):548–53. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 60.Chappell CL, et al. Cryptosporidium hominis: experimental challenge of healthy adults. Am J Trop Med Hyg. 2006;75(5):851–7. [PubMed] [Google Scholar]
- 61.Nash TE, et al. Antigenic variation of Giardia Lamblia in experimental human infections. J Immunol. 1990;144(11):4362–9. [PubMed] [Google Scholar]
- 62.Nash TE, et al. Experimental human infections with Giardia Lamblia. J Infect Dis. 1987;156(6):974–84. doi: 10.1093/infdis/156.6.974. [DOI] [PubMed] [Google Scholar]
- 63.Kotloff KL, et al. A modified Shigella volunteer challenge model in which the inoculum is administered with bicarbonate buffer: clinical experience and implications for Shigella infectivity. Vaccine. 1995;13(16):1488–94. doi: 10.1016/0264-410x(95)00102-7. [DOI] [PubMed] [Google Scholar]
- 64.Erdman DD, Gary GW, Anderson LJ. Serum immunoglobulin a response to Norwalk virus infection. J Clin Microbiol. 1989;27(6):1417–8. doi: 10.1128/jcm.27.6.1417-1418.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Platts-Mills JA, Liu J, Houpt ER. New concepts in diagnostics for infectious diarrhea. Mucosal Immunol. 2013;6(5):876–85. doi: 10.1038/mi.2013.50. [DOI] [PubMed] [Google Scholar]
- 66.Priest JW, et al. Multiplex assay detection of immunoglobulin G antibodies that recognize Giardia intestinalis and Cryptosporidium Parvum antigens. Clin Vaccine Immunol: CVI. 2010;17(11):1695–707. doi: 10.1128/CVI.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Priest JW, et al. Seroepidemiology of Toxoplasma in a coastal region of Haiti: multiplex bead assay detection of immunoglobulin G antibodies that recognize the SAG2A antigen. Epidemiol Infect. 2015;143(3):618–30. doi: 10.1017/S0950268814001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moss DM, et al. Longitudinal evaluation of enteric protozoa in Haitian children by stool exam and multiplex serologic assay. Am J Trop Med Hyg. 2014;90(4):653–60. doi: 10.4269/ajtmh.13-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moss DM, et al. Detection of cryptosporidium antibodies in sera and oral fluids using multiplex bead assay. J Parasitol. 2004;90(2):397–404. doi: 10.1645/GE-3267. [DOI] [PubMed] [Google Scholar]
- 70.Krueger WS, et al. Drinking water source and human Toxoplasma gondii infection in the United States: a cross-sectional analysis of NHANES data. BMC Public Health. 2014;14:711. doi: 10.1186/1471-2458-14-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krueger WS, et al. Environmental risk factors associated with Helicobacter pylori seroprevalence in the United States: a cross-sectional analysis of NHANES data. Epidemiol Infect. 2015;143(12):2520–31. doi: 10.1017/S0950268814003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farkas K, et al. Serological responses to cryptosporidium antigens in inhabitants of Hungary using conventionally filtered surface water and riverbank filtered drinking water. Epidemiol Infect. 2015;143(13):2743–7. doi: 10.1017/S0950268814003859. [DOI] [PubMed] [Google Scholar]
- 73.Frost FJ, et al. Analysis of serological responses to cryptosporidium antigen among NHANES III participants. Ann Epidemiol. 2004;14(7):473–8. doi: 10.1016/j.annepidem.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Kozisek F, et al. Serological responses to cryptosporidium-specific antigens in Czech populations with different water sources. Epidemiol Infect. 2008;136(2):279–86. doi: 10.1017/S0950268807008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tollestrup K, et al. Cryptosporidium infection, onsite wastewater systems and private wells in the arid Southwest. J Water Health. 2014;12(1):161–72. doi: 10.2166/wh.2013.049. [DOI] [PubMed] [Google Scholar]
- 76.Messner M, et al. An approach for developing a national estimate of waterborne disease due to drinking water and a national estimate model application. J Water Health. 2006;4(Suppl 2):201–40. doi: 10.2166/wh.2006.024. [DOI] [PubMed] [Google Scholar]
- 77.McDonald AC, et al. Cryptosporidium Parvum-specific antibody responses among children residing in Milwaukee during the 1993 waterborne outbreak. J Infect Dis. 2001;183(9):1373–9. doi: 10.1086/319862. [DOI] [PubMed] [Google Scholar]
- 78.Mac Kenzie WR, et al. A massive outbreak in Milwaukee of cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331(3):161–7. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 79.Teunis PF, et al. Campylobacter seroconversion rates in selected countries in the European Union. Epidemiol Infect. 2013;141(10):2051–7. doi: 10.1017/S0950268812002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frost F, et al. Serological responses to Cryptosporidium antigens among women using riverbank-filtered water, conventionally filtered surface water and groundwater in Hungary. J Water Health. 2005;3(1):77–82. [PubMed] [Google Scholar]
- 81.Teunis PF, et al. Biomarker dynamics: estimating infection rates from serological data. Stat Med. 2012;31(20):2240–8. doi: 10.1002/sim.5322. [DOI] [PubMed] [Google Scholar]
- 82.Falkenhorst G, et al. Serological cross-sectional studies on salmonella incidence in eight European countries: no correlation with incidence of reported cases. BMC Public Health. 2012;12:523. doi: 10.1186/1471-2458-12-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nogareda F, et al. Incidence and prevalence of Toxoplasma gondii infection in women in France, 1980-2020: model-based estimation. Epidemiol Infect. 2014;142(8):1661–70. doi: 10.1017/S0950268813002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goeyvaerts N, et al. Estimating vaccination coverage for the trivalent measles-mumps-rubella vaccine from trivariate serological data. Stat Med. 2012;31(14):1432–49. doi: 10.1002/sim.4481. [DOI] [PubMed] [Google Scholar]
- 85.Arnold BF, et al. Serological measures of malaria transmission in Haiti: comparison of longitudinal and cross-sectional methods. PLoS One. 2014;9(4):e93684. doi: 10.1371/journal.pone.0093684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nations U. 2030 Agenda: Sustainable Development Goal 6 to ensure availability and sustainable management of water and sanitation for all. 2015 doi: 10.1186/s12889-022-14316-0. Available from: https://sustainabledevelopment.un.org/topics/waterandsanitation. [DOI] [PMC free article] [PubMed]
- 87.Kaminski RW, et al. Multiplexed immunoassay to assess Shigella-specific antibody responses. J Immunol Methods. 2013;393(1–2):18–29. doi: 10.1016/j.jim.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 88.Armah G, et al. A randomized, controlled trial of the impact of alternative dosing schedules on the immune response to human rotavirus vaccine in rural Ghanaian infants. J Infect Dis. 2016 doi: 10.1093/infdis/jiw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qadri F, et al. Peru-15, a live attenuated oral cholera vaccine, is safe and immunogenic in Bangladeshi toddlers and infants. Vaccine. 2007;25(2):231–8. doi: 10.1016/j.vaccine.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 90.Qadri F, et al. Randomized, controlled study of the safety and immunogenicity of Peru-15, a live attenuated oral vaccine candidate for cholera, in adult volunteers in Bangladesh. J Infect Dis. 2005;192(4):573–9. doi: 10.1086/432074. [DOI] [PubMed] [Google Scholar]
- 91.Lemon SM. Immunologic approaches to assessing the response to inactivated hepatitis a vaccine. J Hepatol. 1993;18(Suppl 2):S15–9. doi: 10.1016/s0168-8278(05)80372-1. [DOI] [PubMed] [Google Scholar]
- 92.Zhang J, et al. Protection against hepatitis E virus infection by naturally acquired and vaccine-induced immunity. Clin Microbiol Infect. 2014;20(6):O397–405. doi: 10.1111/1469-0691.12419. [DOI] [PubMed] [Google Scholar]
- 93.Ang CW, et al. Seroepidemiological studies indicate frequent and repeated exposure to campylobacter spp. during childhood. Epidemiol Infect. 2011;139(9):1361–8. doi: 10.1017/S0950268810002359. [DOI] [PubMed] [Google Scholar]
- 94.Mogensen TH, Paludan SR. Molecular pathways in virus-induced cytokine production. Microbiol Mol Biol Rev. 2001;65(1):131–50. doi: 10.1128/MMBR.65.1.131-150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Newman KL, et al. Human norovirus infection and the acute serum cytokine response. Clin Exp Immunol. 2015;182(2):195–203. doi: 10.1111/cei.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brandtzaeg P. Gate-keeper function of the intestinal epithelium. Benefic Microbes. 2013;4(1):67–82. doi: 10.3920/BM2012.0024. [DOI] [PubMed] [Google Scholar]
- 97.Cao AT, et al. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol. 2012;189(9):4666–73. doi: 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Corthesy B. Role of secretory immunoglobulin a and secretory component in the protection of mucosal surfaces. Future Microbiol. 2010;5(5):817–29. doi: 10.2217/fmb.10.39. [DOI] [PubMed] [Google Scholar]
- 99.Riis JL, et al. Salivary cytokines as a minimally-invasive measure of immune functioning in young children: correlates of individual differences and sensitivity to laboratory stress. Dev Psychobiol. 2015;57(2):167. doi: 10.1002/dev.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.JJaedicke KM, Preshaw PM, Taylor JJ. Salivary cytokines as biomarkers of periodontal diseases. Periodontol 2000. 2016;70(1):164–83. doi: 10.1111/prd.12117. [DOI] [PubMed] [Google Scholar]
- 101.Baqui AA, et al. Enhanced interleukin 1 beta, interleukin 6 and tumor necrosis factor alpha in gingival crevicular fluid from periodontal pockets of patients infected with human immunodeficiency virus 1. Oral Microbiol Immunol. 2000;15(2):67–73. doi: 10.1034/j.1399-302x.2000.150201.x. [DOI] [PubMed] [Google Scholar]
- 102.Desai GS, Mathews ST. Saliva as a non-invasive diagnostic tool for inflammation and insulin-resistance. World J Diabetes. 2014;5(6):730–8. doi: 10.4239/wjd.v5.i6.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spear GT, et al. Relationship of HIV RNA and cytokines in saliva from HIV-infected individuals. FEMS Immunol Med Microbiol. 2005;45(2):129–36. doi: 10.1016/j.femsim.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 104.Aleksandra Nielsen A, et al. Saliva interleukin-6 in patients with inflammatory bowel disease. Scand J Gastroenterol. 2005;40(12):1444–8. doi: 10.1080/00365520510023774. [DOI] [PubMed] [Google Scholar]
- 105.Kaslow RA, Evans AS. Epidemiologic concepts and methods. In: Evans AS, Kaslow RA, editors. Viral infections of humans: epidemiology and control. Plenum Publishing Corporation; New York, NY: 1997. pp. 3–58. [Google Scholar]
- 106.Priest JW, et al. Enzyme immunoassay detection of antigen-specific immunoglobulin g antibodies in longitudinal serum samples from patients with cryptosporidiosis. Clin Diagn Lab Immunol. 2001;8(2):415–23. doi: 10.1128/CDLI.8.2.415-423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crump JA, et al. Comparing serologic response against enteric pathogens with reported diarrhea to assess the impact of improved household drinking water quality. Am J Trop Med Hyg. 2007;77(1):136–41. [PubMed] [Google Scholar]
- 108.Sarkar R, et al. Serum IgG responses and seroconversion patterns to Cryptosporidium gp15 among children in a birth cohort in South India. Clin Vaccine Immunol : CVI. 2012;19(6):849–54. doi: 10.1128/CVI.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Becker DJ, Oloya J, Ezeamama AE. Household socioeconomic and demographic correlates of Cryptosporidium seropositivity in the United States. PLoS Negl Trop Dis. 2015;9(9):e0004080. doi: 10.1371/journal.pntd.0004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Checkley W, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis. 2015;15(1):85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cozon G, et al. Secretory IgA antibodies to Cryptosporidium parvum in AIDS patients with chronic cryptosporidiosis. J Infect Dis. 1994;169(3):696–9. doi: 10.1093/infdis/169.3.696. [DOI] [PubMed] [Google Scholar]
- 112.Egorov AI, et al. Recent diarrhea is associated with elevated salivary IgG responses to Cryptosporidium in residents of an eastern Massachusetts community. Infection. 2010;38(2):117–23. doi: 10.1007/s15010-009-9323-4. [DOI] [PubMed] [Google Scholar]
- 113.Rokosz-Chudziak N, Rastawicki W. Frequency of antibodies to the recombinant protein P39 of C. jejuni in patients with gastrointestinal disorders and reactive arthritis in Poland. Med Dosw Mikrobiol. 2014;66(3–4):195–207. [PubMed] [Google Scholar]
- 114.Tribble DR, et al. Assessment of the duration of protection in campylobacter jejuni experimental infection in humans. Infect Immun. 2010;78(4):1750–9. doi: 10.1128/IAI.01021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Islam D, et al. Immune responses to Campylobacter (C. jejuni or C. coli) infections: a two-year study of US forces deployed to Thailand. APMIS. 2014;122(11):1102–13. doi: 10.1111/apm.12266. [DOI] [PubMed] [Google Scholar]
- 116.Cawthraw SA, et al. Long-term antibody responses following human infection with campylobacter jejuni. Clin Exp Immunol. 2002;130(1):101–6. doi: 10.1046/j.1365-2249.2002.01966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiménez JC, et al. Antibody response in children infected with Giardia Intestinalis before and after treatment with Secnidazole. Am J Trop Med Hyg. 2009;80(1):11–5. [PubMed] [Google Scholar]
- 118.Rodriguez OL, et al. Secretory IgA antibody responses in Venezuelan children infected with Giardia duodenalis. J Trop Pediatr. 2004;50(2):68–72. doi: 10.1093/tropej/50.2.68. [DOI] [PubMed] [Google Scholar]
- 119.El-Gebaly NS, et al. Saliva and sera IgA and IgG in Egyptian Giardia-infected children. Parasitol Res. 2012;111(2):571–5. doi: 10.1007/s00436-012-2869-y. [DOI] [PubMed] [Google Scholar]
- 120.Hundekar S, et al. Viral excretion and antibody titers in children infected with hepatitis a virus from an orphanage in western India. J Clin Virol. 2015;73:27–31. doi: 10.1016/j.jcv.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 121.Laufer DS, et al. Saliva and serum as diagnostic media for antibody to hepatitis a virus in adults and in individuals who have received an inactivated hepatitis a vaccine. Clin Infect Dis. 1995;20(4):868–71. doi: 10.1093/clinids/20.4.868. [DOI] [PubMed] [Google Scholar]
- 122.Ochnio JJ, et al. New, ultrasensitive enzyme immunoassay for detecting vaccine- and disease-induced hepatitis A virus-specific immunoglobulin G in saliva. J Clin Microbiol. 1997;35(1):98–101. doi: 10.1128/jcm.35.1.98-101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tourinho RS, et al. Cross-sectional study of hepatitis A virus infection in the Pantanal population before vaccine implementation in Brazil: usage of non-invasive specimen collection. Int J Environ Res Public Health. 2015;12(7):7357–69. doi: 10.3390/ijerph120707357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Adjei AA, et al. Hepatitis E virus infection is highly prevalent among pregnant women in Accra Ghana. Virol J. 2009;6:108. doi: 10.1186/1743-422X-6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pas SD, et al. Diagnostic performance of selected commercial HEV IgM and IgG ELISAs for immunocompromised and immunocompetent patients. J Clin Virol. 2013;58(4):629–34. doi: 10.1016/j.jcv.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 126.Kumar A, et al. Association of cytokines in hepatitis E with pregnancy outcome. Cytokine. 2014;65(1):95–104. doi: 10.1016/j.cyto.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 127.Gu G, et al. Hepatitis E virus seroprevalence in pregnant women in Jiangsu, China, and postpartum evolution during six years. BMC Infect Dis. 2015;15(1):560. doi: 10.1186/s12879-015-1308-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Heaney CD, et al. Arsenic exposure and hepatitis E virus infection during pregnancy. Environ Res. 2015;142:273–80. doi: 10.1016/j.envres.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kmush BL, et al. The association of cytokines and micronutrients with hepatitis E virus infection during pregnancy and the postpartum period in rural Bangladesh. Am J Trop Med Hyg. 2016;94(1):203–11. doi: 10.4269/ajtmh.15-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Iritani N, et al. Humoral immune responses against norovirus infections of children. J Med Virol. 2007;79(8):1187–93. doi: 10.1002/jmv.20897. [DOI] [PubMed] [Google Scholar]
- 131.Ramani S, et al. Mucosal and cellular immune responses to Norwalk virus. J Infect Dis. 2015;212(3):397–405. doi: 10.1093/infdis/jiv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Grimwood K, et al. Comparison of serum and mucosal antibody responses following severe acute rotavirus gastroenteritis in young children. J Clin Microbiol. 1988;26(4):732–8. doi: 10.1128/jcm.26.4.732-738.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Azim T, et al. Rotavirus-specific subclass antibody and cytokine responses in Bangladeshi children with rotavirus diarrhoea. J Med Virol. 2003;69(2):286–95. doi: 10.1002/jmv.10280. [DOI] [PubMed] [Google Scholar]
- 134.Xu J, et al. Serum antibody responses in children with rotavirus diarrhea can serve as proxy for protection. Clin Diagn Lab Immunol. 2005;12(2):273–9. doi: 10.1128/CDLI.12.2.273-279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Premkumar P, et al. Association of serum antibodies with protection against rotavirus infection and disease in south Indian children. Vaccine. 2014;32(Supplement 1):A55–61. doi: 10.1016/j.vaccine.2014.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sindhu KN, et al. Immune response and intestinal permeability in children with acute gastroenteritis treated with Lactobacillus rhamnosus GG: a randomized, double-blind, placebo-controlled trial. Clin Infect Dis. 2014;58(8):1107–15. doi: 10.1093/cid/ciu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moon SS, et al. Prevaccination rotavirus serum IgG and IgA are associated with lower immunogenicity of live, oral human rotavirus vaccine in south African infants. Clin Infect Dis. 2016;62(2):157–65. doi: 10.1093/cid/civ828. [DOI] [PubMed] [Google Scholar]
- 138.Stals F, Walther FJ, Bruggeman CA. Faecal and pharyngeal shedding of rotavirus and rotavirus IgA in children with diarrhoea. J Med Virol. 1984;14(4):333–9. doi: 10.1002/jmv.1890140406. [DOI] [PubMed] [Google Scholar]
- 139.Aiyar J, et al. Rotavirus-specific antibody response in saliva of infants with rotavirus diarrhea. J Infect Dis. 1990;162(6):1383–4. doi: 10.1093/infdis/162.6.1383. [DOI] [PubMed] [Google Scholar]
- 140.Friedman MG, et al. Subclasses of IgA antibodies in serum and saliva samples of newborns and infants immunized against rotavirus. Clin Exp Immunol. 1996;103(2):206–11. doi: 10.1046/j.1365-2249.1996.d01-620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ludwig K, et al. Antibody response to Shiga toxins Stx2 and Stx1 in children with enteropathic hemolytic-uremic syndrome. J Clin Microbiol. 2001;39(6):2272–9. doi: 10.1128/JCM.39.6.2272-2279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kulkarni H, et al. Escherichia coli ‘O’ group serological responses and clinical correlations in epidemic HUS patients. Comp Immunol Microbiol Infect Dis. 2002;25(4):249–68. doi: 10.1016/s0147-9571(02)00011-5. [DOI] [PubMed] [Google Scholar]
- 143.Fernández-Brando RJ, et al. Antibody response to Shiga toxins in Argentinean children with Enteropathic hemolytic uremic syndrome at acute and long-term follow-up periods. PLoS One. 2011;6(4):1–7. doi: 10.1371/journal.pone.0019136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guirro M, et al. Humoral immune response to Shiga toxin 2 (Stx2) among Brazilian urban children with hemolytic uremic syndrome and healthy controls. BMC Infect Dis. 2014;14:320. doi: 10.1186/1471-2334-14-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ludwig K, et al. Saliva IgM and IgA are a sensitive indicator of the humoral immune response to Escherichia coli O157 lipopolysaccharide in children with enteropathic hemolytic uremic syndrome. Pediatr Res. 2002;52(2):307–13. doi: 10.1203/00006450-200208000-00026. [DOI] [PubMed] [Google Scholar]
- 146.Chart H, et al. Analysis of saliva for antibodies to the LIPS of Escherichia coli O157 in patients with serum antibodies to E-coli O157 LIPS. J Med Microbiol. 2003;52(7):569–72. doi: 10.1099/jmm.0.05126-0. [DOI] [PubMed] [Google Scholar]
- 147.Van De Verg LL, et al. Cross-reactivity of Shigella flexneri serotype 2a O antigen antibodies following immunization or infection. Vaccine. 1996;14(11):1062–8. doi: 10.1016/0264-410x(96)00006-0. [DOI] [PubMed] [Google Scholar]
- 148.Raqib R, et al. A systemic downregulation of gamma interferon production is associated with acute shigellosis. Infect Immun. 1997;65(12):5338–41. doi: 10.1128/iai.65.12.5338-5341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rasolofo-Razanamparany V, et al. Predominance of serotype-specific mucosal antibody response in Shigella flexneri-infected humans living in an area of endemicity. Infect Immun. 2001;69(9):5230–4. doi: 10.1128/IAI.69.9.5230-5234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Levine MM, et al. Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road. Nat Rev Microbiol. 2007;5(7):540–53. doi: 10.1038/nrmicro1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Muhsen K, et al. Age-dependent association among helicobacter pylori infection, serum pepsinogen levels and immune response of children to live oral cholera vaccine CVD 103-HgR. PLoS One. 2014;9(1):e83999. doi: 10.1371/journal.pone.0083999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Thompson CN, et al. A cohort study to define the age-specific incidence and risk factors of Shigella diarrhoeal infections in Vietnamese children: a study protocol. BMC Public Health. 2014;14:1289. doi: 10.1186/1471-2458-14-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Azim T, et al. Cytokines in the stools of children with complicated shigellosis. Clin Diagn Lab Immunol. 1995;2(4):492–5. doi: 10.1128/cdli.2.4.492-495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schultsz C, et al. Shigella-specific IgA in saliva of children with bacillary dysentery. FEMS Microbiol Immunol. 1992;4(2):65–72. doi: 10.1111/j.1574-6968.1992.tb04972.x. [DOI] [PubMed] [Google Scholar]
- 155.Chowdhury F, et al. A comparison of clinical and immunologic features in children and older patients hospitalized with severe cholera in Bangladesh. Pediatr Infect Dis J. 2008;27(11):986–92. doi: 10.1097/INF.0b013e3181783adf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Johnson RA, et al. Comparison of immune responses to the O-specific polysaccharide and lipopolysaccharide of Vibrio cholerae O1 in Bangladeshi adult patients with cholera. Clin Vaccine Immunol: CVI. 2012;19(11):1712–21. doi: 10.1128/CVI.00321-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fujii Y, et al. Serological surveillance development for tropical infectious diseases using simultaneous microsphere-based multiplex assays and finite mixture models. PLoS Negl Trop Dis. 2014;8(7):1–15. doi: 10.1371/journal.pntd.0003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Khan AI, et al. Cholera in pregnancy: clinical and immunological aspects. Int J Infect Dis. 2015;39:20–4. doi: 10.1016/j.ijid.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Qadri F, et al. Antigen-specific immunoglobulin A antibodies secreted from circulating B cells are an effective marker for recent local immune responses in patients with cholera: comparison to antibody-secreting cell responses and other immunological markers. Infect Immun. 2003;71(8):4808–14. doi: 10.1128/IAI.71.8.4808-4814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jertborn M, Svennerholm AM, Holmgren J. Saliva, breast milk, and serum antibody responses as indirect measures of intestinal immunity after oral cholera vaccination or natural disease. J Clin Microbiol. 1986;24(2):203–9. doi: 10.1128/jcm.24.2.203-209.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]


