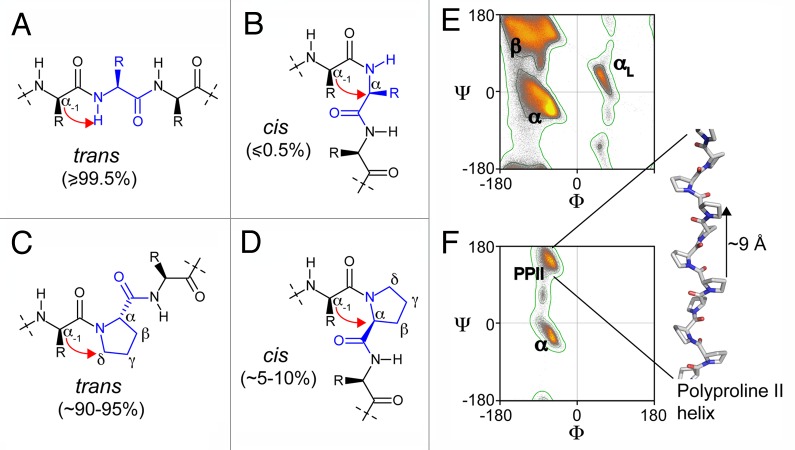
Figure 2. Chemical structure of peptide fragments in trans (A) and (C) and cis conformation (B) and (D); (C) and (D) show a proline-containing fragment. The red arrows point out the steric hindrances between the Cα of the residue (−1) with the Hamide (A) or the Cα of the residue (0) (B) for the non-proline-containing peptides, and between the Cα of the residue (-1) with the Cδ (C) or the Cα of the proline (D). Ramachandran plots of non-proline, non-glycine, non-isoleucine, non-valine residues (E) and proline residues (F) result from the analysis of 1.5 million residues in 8,000 protein chains with resolution < 2 Å and backbone B-factors < 30.The contours separate the “outlier,” “allowed” and “favored” regions of the Ramachandran plots. The Ramachandran plots were adapted from commons.wikimedia.org/wiki/User:Dcrjsr. The β-strand (β), α-helix (α), α-L-helix (αL), poly-proline II (PPII) regions of the Ramachandran plots are indicated and we show a representation of a model poly-proline II helix.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.