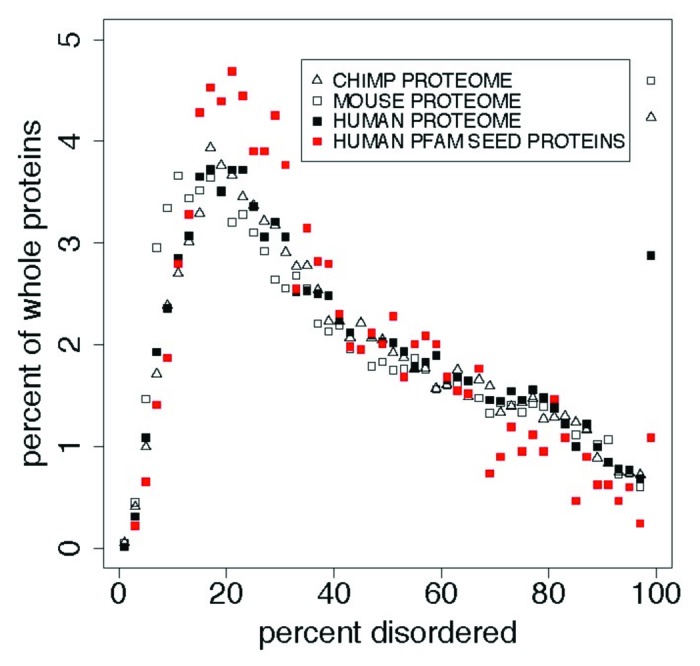
Figure 2. In whole proteins there appears to be a predominance of 100% PID over what is found in the Pfam seed database for humans, with more (here) in mouse and chimp than in human proteins. This cannot be attributed to an uneven distribution of Pfam domain or PID lengths, and suggests that the Pfam seed database excludes some IDPs. This is shown here in comparisons of the distribution of percent predicted disorder in the human proteome with the PanTroglodytes chimpanzee and MusMusculus mouse proteomes, and with human proteins chosen as sources for Pfam seed members (red). Table 3 shows statistics for these distributions. Proteins with 0% disorder are not plotted here. We note that the distribution for mouse is shifted 1% to the left of that for chimp and human proteomes while 100% PID is highest for the mouse.
