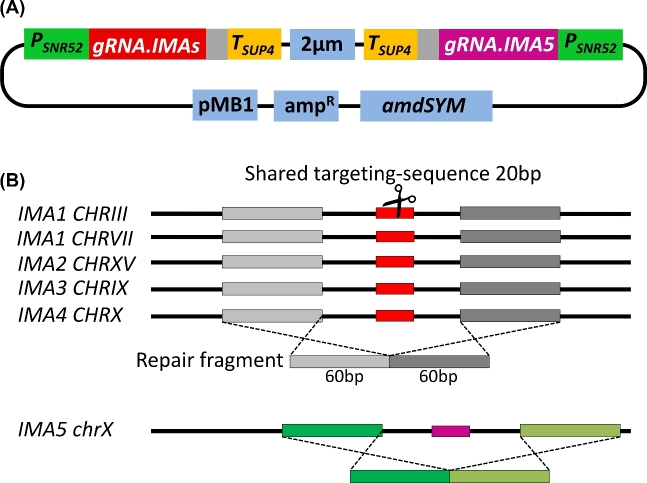
Figure 1.
Strategy for deletion of the IMA genes using a shared Cas9 target site. (A) Plasmid pUDR127, that contains two gRNA-cassettes: ‘gRNA-IMAs’ (in red) with a target sequence to cleave all IMA genes except IMA5 and ‘gRNA-IMA5’, in purple. The SNR52 promoter and SUP4 terminator are shown in green and yellow, respectively. Sequences encoding structural RNAs are shown in grey. 2 μm: origin of replication in yeast; pMB1: origin of replication in bacteria; ampR: cassette for ampicillin resistance in E. coli; amdSYM: dominant marker that allows use of acetamide as nitrogen source. (B) Illustration of the shared Cas9 target sequences among the IMA genes. The coding sequence of each IMA is represented by black lines. The red boxes correspond to the shared target sequence. The dark- and light-grey fragments are the homology regions (60 bp) up- and downstream of the target site where the repair fragment (120 bp) can recombine. In view of its sequence divergence from the other IMA genes, an exclusive target sequence, in purple, and repair sites, in dark and light green, were chosen for inactivation of IMA5.