Figure 5. Figure 5: Interactions at the interface side of cytoplasmic pre-60S ribosomal particles.
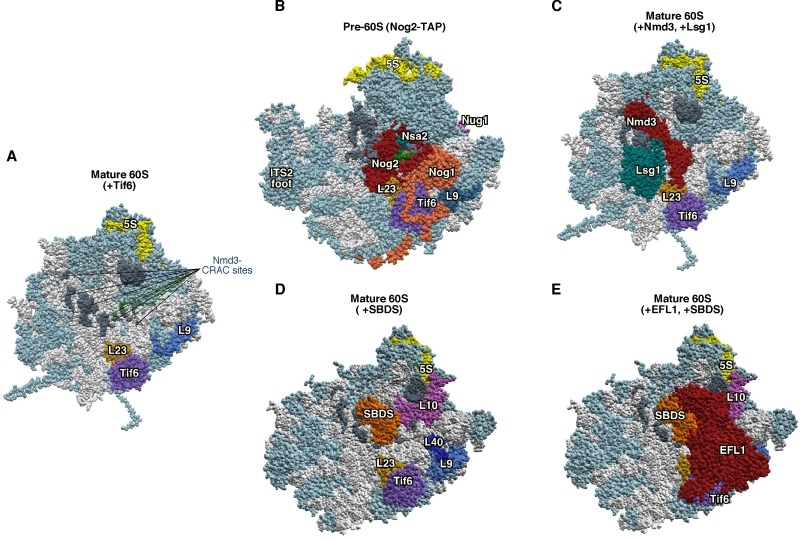
(A) The Nmd3-binding sites, identified by CRAC at 25S rRNA helices H38, H69-69 and H89-90 115, have been highlighted in dark grey in a reconstituted 60S r-subunit (PDB ID:5T62; 114). The CRAC sites common for Nmd3, Dbp10 and Nug1 115,118 are labelled in green.
(B) Position of Nog2 (red), Nsa2 (blue), Nog1 (gold) and Nug1 (pink) in the early pre-60S r-particle purified with Nog2-TAP (PDB ID: 3JCT; 40). Note that only a very small portion of Nug1 has been resolved in this particle.
(C) Position of Nmd3 and Lsg1 in a reconstituted 60S r-subunit (PDB ID:5T62; 114). Note that only the region of Nmd3 comprised between residues 46 and 388 of 518 in total is shown. The N-terminal end of Nmd3 approaches to Tif6 while the C-terminal end contact L1. In A-C, the position of Tif6 (purple) is shown. The locations of the 5S rRNA (yellow) and that of the L23 (gold) and L9 r-proteins (royal blue) have also been highlighted.
(D) Position of SBDS (dark gold) in the 60S r-subunit. The structure of a pre-60S r-particle from Dyctiostelium discoideum containing Tif6 and reconstituted with human SBDS and EFL1 (PDB ID: 5ANB; 116) was superimposed on the structure of yeast 60S r-subunit (PDB ID: 5APN; 68); then, all common proteins from D. discoideium were removed from the model. L9 (royal blue), L40 (navy blue), L10 (violet), L23 (gold) and 5S rRNA (yellow) were highlighted. Note that SBDS is shown in its open conformation state.
(E) As D, but additionally showing EFL1 (red) on top of Tif6. In all figures, the Nmd3-CRAC sites shown in A (dark grey) were also highlighted, the rest of rRNAs are coloured in pale blue and the rest of r-proteins and/or factors in light cornflower blue.
