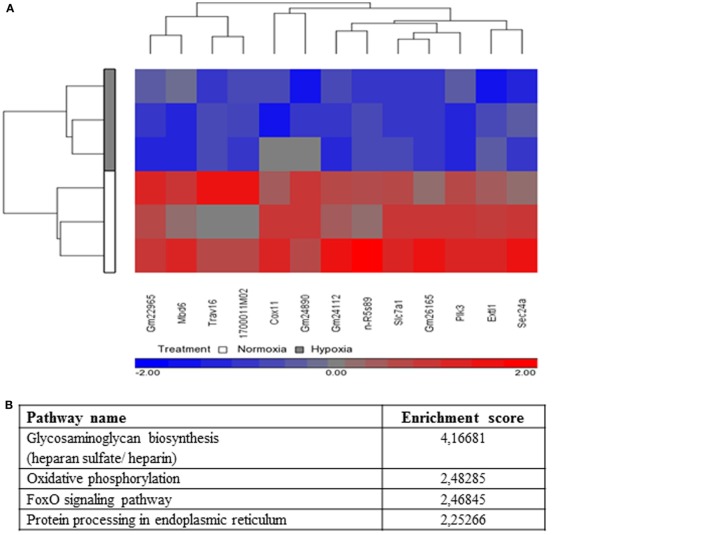
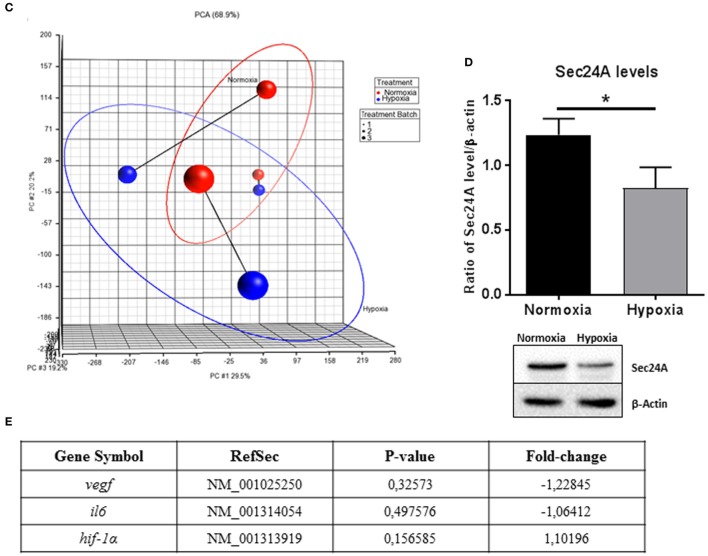
Figure 7.
Microarray expression analysis. (A) Hierarchical clustering of genes with significantly different expression across hypoxia versus normoxia (change in hypoxia relative to normoxia with p-value <0.05, fold change > 1.3, or fold change < −1.3). Genes that are upregulated appear in red, and those that are downregulated appear in blue. (B) Selected top pathways from the Partek analysis sorted by the Enrichment Score (Fisher’s Exact test). (C) Principle component analysis (PCA) of microarray results (n = 3). The blue (hypoxic-treated cells) and red (normoxic-treated cells) oval dots represent linear combinations of the expression data, including relative expression value and variance. The PCA in PGS software examines three components of genes in different samples for those with similar or different expression profiles. PCA (Partek software) was used to compare the gene expression signatures of hypoxia with those of normoxia and connected to treatment Batch (1, 2, and 3). The percentage of overall variance is indicated for each PC and for the combination of all three PCs. The two ellipses show how the samples are grouped by treatment and line-connected by treatment batch. (D) Sec24A protein levels of whole cell lysates of mast cells (n = 3) after 3 h under hypoxia and normoxia. Sec24A was normalized to β-actin (ratio of Sec24A/β-actin). Significantly less Sec24A protein was detected after 3 h under hypoxia compared to normoxia. Differences between the two groups were analyzed by using a paired, two-tailed t-test. Representative western blot: total cell extracts (50 μg) were separated by SDS/PAGE (10% gel) and transferred onto PVDF membranes (Roth, Germany). Sec24A protein (66 kDa) was detected by the primary antibody against Sec24A (rabbit anti-Sec24A, Proteintech, 0.2 μg/ml). β-actin (43 kDa) was used as a housekeeping gene (mouse anti-β actin, Santa Cruz). (E) Microarray transcript expression analysis of hif-1α and its target genes vegf and il-6. No significant changes in gene expression are observable.