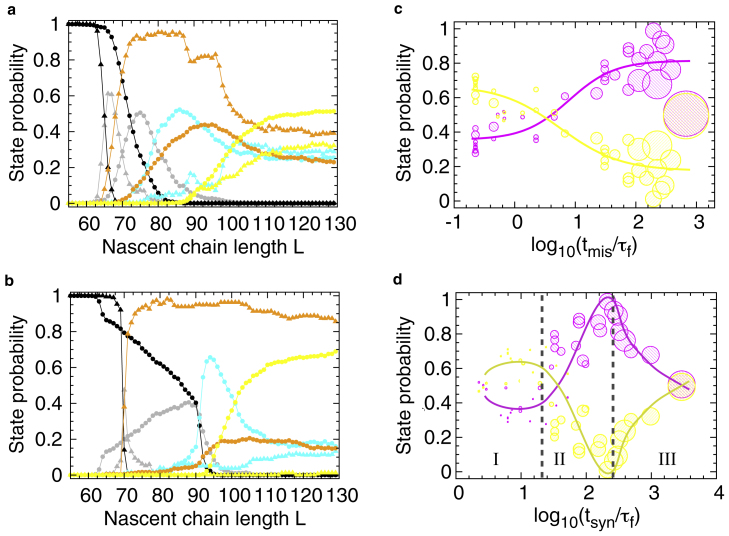
Figure 4.
The effect of different elongation schedules on cotranslational folding and misfolding. The probabilities of populating states U (black), D (gray), M (orange), C (cyan), and N (yellow) as a function of nascent chain length L, when one amino acid is added every τaa(L) ns (elongation schedules s1 to s40 are reported in Fig. S4 c). In all panels, lines are meant as a guide to the eye and the standard errors about the mean are smaller than the symbols. (a) State probabilities from the uniformly slow (s30) and uniformly fast (s2) elongation schedules are shown, respectively, as triangles and circles. Each data point is an average over 300 independent synthesis trajectories. (b) Same as (a), except the state probabilities resulting from the nonuniform, slow schedule s39 (triangles) and nonuniform, fast schedule s5 (circles) are shown. (c) Shown here are the native (yellow) and nonnative (M and C; magenta) state probabilities at the end of synthesis (i.e., at L = 129 residues) as a function of the reduced time Tmis = tmis/τf. The value tmis is the time necessary to synthesize residues 70–90 and τf = 1.0 ns is the mean folding time of the fast-kinetic phase of this protein off the ribosome. The data point sizes are proportional to the reduced total time Tsyn = tsyn/τf needed to synthesize the entire protein (from residue 40–129). Each data point results from one of the elongation schedules shown in Fig. S4 c. (d) Same as (c), but as a function of the reduced time Tsyn = tsyn/τf. The value tsyn is the time necessary to synthesize the entire protein from residues 40–129. Circle sizes are proportional to the time Tmis needed to synthesize the misfolding-prone segment. Vertical dashed lines indicate boundaries among regions I, II, and III.