Figure 6.
Epigenetic Modifications Induced by Nuclear Reprogramming Lead to Therapy Resistance against MAPK Inhibitors in Melanoma Cells
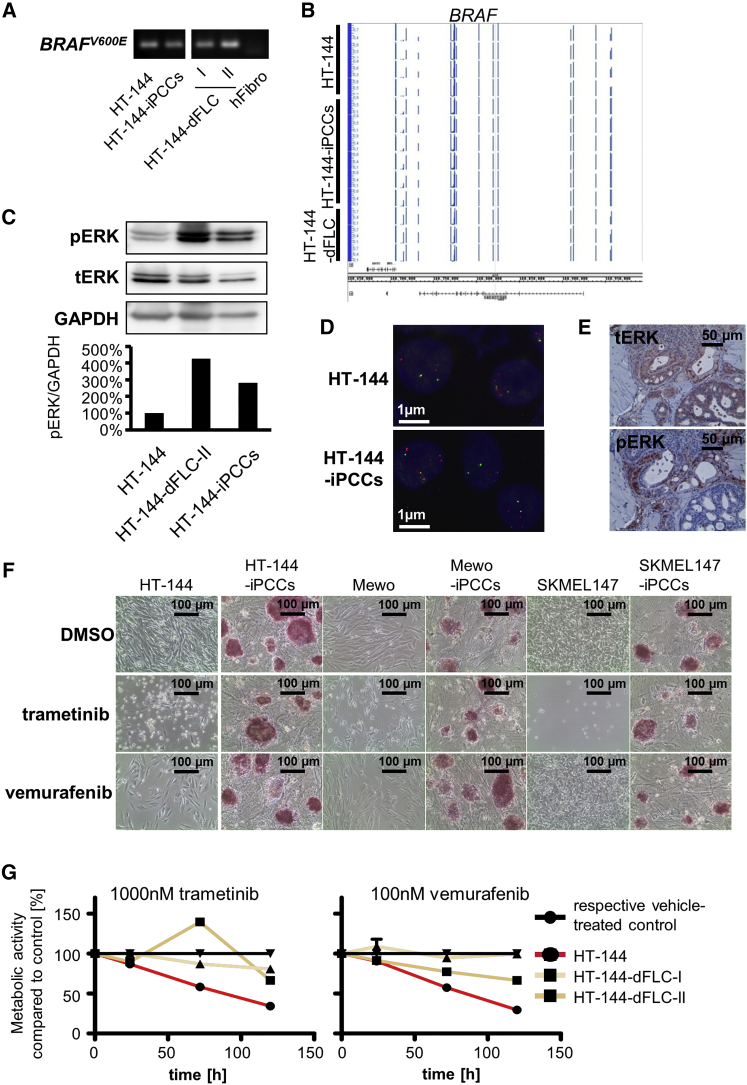
(A) Oncogene-specific PCR analysis of BRAFV600E in the parental HT-144 cells, HT-144-iPCCs, and iPCC-derived fibroblast-like cells. Human fibroblasts were used as a negative control.
(B) Methylation analysis of the BRAF locus in HT-144, HT-144-iPCCs, and HT-144-dFLCs.
(C) ERK activity in reprogrammed HT-144-iPCCs and fibroblast-like cells compared with the parental cell line determined by western blot analysis of phosphorylated ERK. GAPDH was used as an internal reference control for semi-quantitative protein analysis.
(D) FISH of HT144 and HT144-iPCCs shows no gain or loss of gene copy numbers related to genes of the BRAF-MEK-ERK signaling pathway such as BRAF (shown are nuclei with signals for the BRAF gene locus [red] and a centromeric reference probe [green]).
(E) HT-144-iPCCs give rise to tumors with reduced ERK activity in distinct differentiated structures. Histological staining of total ERK (tERK) and phosphorylated ERK (pERK) in HT-144-iPCC-derived tumors.
(F) Treatment of parental melanoma cells and their reprogrammed counterparts with the MEK inhibitor trametinib and the BRAFV600E-specific inhibitor vemurafenib.
(G) Therapy response of MAPK inhibitor-treated HT-144 and two fibroblast-like in vitro differentiations. Cells were incubated with 1,000 nM trametinib and 100 nM vemurafenib and analyzed for their metabolic activity at indicated time points.
See also Figure S5.