Summary
The ability to form teratomas in vivo containing multiple somatic cell types is regarded as functional evidence of pluripotency for human pluripotent stem cells (hPSCs). Since the Teratoma assay is animal dependent, laborious, and only qualitative, the PluriTest and the hPSC ScoreCard assay have been developed as in vitro alternatives. Here we compared normal hPSCs, induced hPSCs (hiPSCs) with reactivated reprogramming transgenes, and human embryonal carcinoma cells (hECs) in these assays. While normal hPSCs gave rise to typical teratomas, the xenografts of the hECs and the hiPSCs with reactivated reprogramming transgenes were largely undifferentiated and malignant. The hPSC ScoreCard assay confirmed the line-specific differentiation propensities in vitro. However, when undifferentiated cells were analyzed by the PluriTest, only hECs were identified as abnormal whereas all other cell lines were indistinguishable and resembled normal hPSCs. Our results indicate that pluripotency assays are best selected on the basis of intended downstream applications.
Keywords: human pluripotent stem cell, pluripotency assay, teratoma, PluriTest, hPSC ScoreCard, TeratoScore, tumorigenicity, malignancy, differentiation-defective
Highlights
-
•
Side-by-side comparison of teratomas/TeratoScore, hPSC ScoreCard, and PluriTest
-
•
hiPSCs with reactivated transgenes form embryonal carcinomas in vivo
-
•
hiPSCs with reactivated transgenes show impaired differentiation capacity in vitro
• PluriTest does not distinguish hiPSCs with reactivated transgenes from normal hPSCs
Salvatori, Freund and colleagues performed a side-by-side comparison of Teratoma assay, ScoreCard, and PluriTest using normal hPSCs, hiPSCs with reactivated reprogramming transgenes, and embryonal carcinoma cells. For assessment of pluripotency the Teratoma assay can be replaced by a combination of ScoreCard and PluriTest. Notably, only the Teratoma assay is able to reveal the potential malignancy of hPSCs.
Introduction
Since the advent of cellular reprogramming with exogenous transcription factors (Takahashi et al., 2007, Takahashi and Yamanaka, 2006), human induced pluripotent stem cells (hiPSCs) have demonstrated important potential for research on differentiation in human development, modeling congenital diseases, drug target identification, and safety pharmacology (Passier et al., 2016). hiPSC-derived differentiated cells are also expected to play an increasing role in human cell therapy (Inoue et al., 2014).
For optimal use, it is essential to identify hiPSC lines that are fully reprogrammed and of high quality with proven pluripotency in terms of differentiation to derivatives of three germ layers. Parameters identified as affecting differentiation include the genetic background (Choi et al., 2015, Kyttala et al., 2016), X-inactivation status in female lines (Anguera et al., 2012), the reprogramming vector used (Choi et al., 2015), the combination of the reprogramming factors (Buganim et al., 2014), their stoichiometry (Carey et al., 2011), or their incomplete silencing after reprogramming (Ohnuki et al., 2014). A simple assay to determine their differentiation capacity prospectively would significantly improve the efficiency of hiPSC selection for further use.
At the molecular level, the pluripotency status is defined by a set of commonly expressed marker genes (International Stem Cell Initiative et al., 2007) as well as epigenetic features such as demethylated pluripotency gene promoters and the presence of bivalent domains in developmental gene regions (Maherali and Hochedlinger, 2008). Currently there is no clear consensus on the minimal requirements that constitute pluripotency at the molecular level.
Functional pluripotency, on the other hand, is defined as the ability to form differentiated cell types of the three germ layers. Whereas mouse PSCs are tested for their ability to contribute to chimeric embryos or to form the entire organism in vivo, the “Teratoma assay” has been developed as a surrogate for functional pluripotency in human stem cells (Daley et al., 2009, International Stem Cell Banking Initiative, 2009). Undifferentiated human pluripotent stem cells (hPSCs) are injected into adult immunocompromised mice, where they form ideally benign-appearing tumor masses containing derivatives of the three germ layers (Gertow et al., 2007). However, the Teratoma assay requires mice, is costly and time consuming, and requires an experienced pathologist for analysis. The biggest drawback is often the lack of quantification of differentiation.
An ongoing debate is whether the Teratoma assay is an acceptable tool to evaluate pluripotency (Buta et al., 2013, Dolgin, 2010). This has led to the search for animal-independent in vitro alternatives as well as suggestions of how to improve the original assay. A recently developed microarray-based algorithm called TeratoScore quantifies the extent to which the query sample resembles a teratoma or a primary tumor (Avior et al., 2015). The hPSC ScoreCard assay quantifies the ability of a hPSC line to differentiate into the three germ layers in vitro (Bock et al., 2011, Tsankov et al., 2015). By contrast, the PluriTest algorithm compares the global gene expression patterns of undifferentiated hPSCs with those of a reference pool consisting of numerous validated hPSCs and differentiated cells (Muller et al., 2011).
Here we compared the outcome of these various pluripotency assays using validated human embryonic stem cells (hESCs), tetraploid hPSCs with a reported mesendodermal differentiation bias, normal hiPSCs, hiPSCs with reactivated (doxycycline [Dox]-inducible) reprogramming factors, and human embryonal carcinoma cells (hECs). We found that hESCs, tetraploid hPSCs, and normal hiPSCs all gave rise to typical teratomas. By contrast, tumors generated from hECs and hiPSCs with reactivated reprogramming factors were largely undifferentiated and malignant. These differences were confirmed by the TeratoScore. However, the algorithm was unable to identify partially differentiated tumors. Short-term in vitro differentiation analyzed by the hPSC ScoreCard assay confirmed that the differentiation of hiPSCs with reactivated transgenes was severely compromised. However, in the PluriTest algorithm, normal hiPSCs and differentiation-defective hiPSCs were indistinguishable. Our data suggest that in vivo and in vitro assays can reveal distinct features of hPSCs (molecular or functional pluripotency, malignancy) and that the choice of the assay(s) depends on the downstream application of a particular hiPSC line.
Results
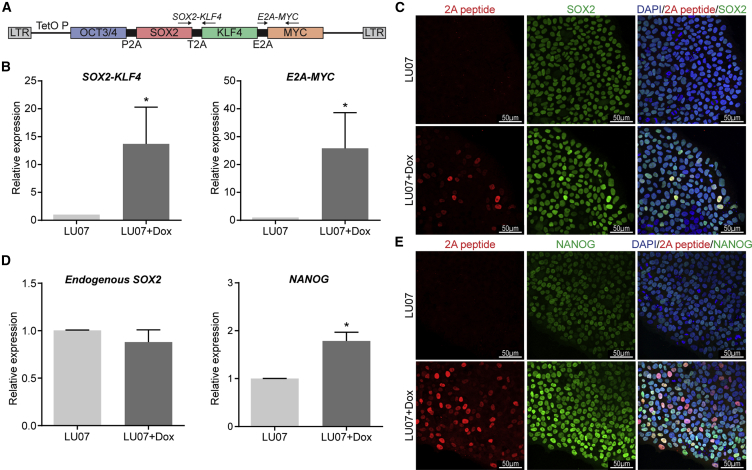
To evaluate and compare the performance of the standard Teratoma assay and the in vitro/in silico pluripotency assays, we selected cell lines which express typical markers of hPSCs but are expected to vary in their ability to differentiate. As a standard line, we used H9 hESCs (H9) (Thomson et al., 1998). Secondly, a tetraploid hybrid line generated by fusion of H9 hESCs and hematopoietic stem cells with a reported differentiation bias toward mesendoderm was used (H9Hyb) (Qin et al., 2014). Thirdly, we generated hiPSCs (LU07) from skin fibroblasts using a polycistronic lentivirus with Dox-inducible transgenes OCT3/4, SOX2, KLF4, and c-MYC (Figure 1A; Carey et al., 2009). LU07 hiPSCs are normally Dox independent and differentiate efficiently in vitro into derivatives of all three germ layers in the presence of fetal calf serum (FCS) (data not shown). However, in the presence of Dox (LU07+Dox), the polycistronic transgene cassette is reactivated, as evidenced by qPCR for exogenous c-MYC, KLF4, and SOX2 (Figure 1B). Immunofluorescent (IF) staining of the transgenic self-cleaving 2A peptide revealed that its levels vary between individual cells and that induction of the 2A peptide leads to an increase in SOX2 protein (Figure 1C). Endogenous SOX2 expression levels were unaltered (Figure 1D), whereas endogenous NANOG was upregulated in LU07+Dox cells (Figures 1D and 1E). Finally we used an hEC line, which expresses pluripotency markers but lacks the ability to differentiate and is therefore considered nullipotent (Josephson et al., 2007). hPSCs were cultured under defined conditions on vitronectin in TESR-E8 medium whereas hECs were maintained in the presence of FCS as described by Josephson et al. (2007). For all assays we used undifferentiated cell populations with ≥85% OCT3/4-expressing cells as determined by fluorescence-activated cell sorting (FACS) (data not shown).
Figure 1.
Generation and Characterization of LU07 hiPSCs with Dox-Inducible Transgenes
(A) Schematic of the lentiviral construct with Dox-inducible reprogramming factors used for the generation of LU07 hiPSCs (Carey et al., 2009). Primers for the detection of transgenic SOX2, KLF4, and c-MYC are indicated by arrows. The 2A peptide (P2A) is located between OCT3/4 and SOX2.
(B) Expression levels of transgenic SOX2, KLF4, and c-MYC in undifferentiated LU07 or LU07 cells treated with Dox for 3 days as determined by qPCR. Average data ± SEM (n = 4 independent experiments). ∗p < 0.05.
(C) Representative IF staining of undifferentiated LU07 and LU07+Dox for 2A peptide and SOX2. Nuclei were stained with DAPI.
(D) Expression levels of endogenous SOX2 and NANOG in undifferentiated LU07 or LU07 cells treated with Dox for 3 days as determined by qPCR. Average data ± SEM (n = 4 independent experiments). ∗p < 0.05.
(E) Representative IF staining of undifferentiated LU07 and LU07+Dox for 2A peptide and NANOG. Nuclei were stained with DAPI.
See also Figure S1.
Furthermore, we tested the genetic integrity with the COBRA assay (Szuhai and Tanke, 2006) in a fraction of cells used for teratoma formation and for PluriTest. As expected, hECs displayed various aneuploidies including additional copies of (partial) chromosomes 1, 12, and 20 (Figure S1). H9Hyb cells were tetraploid and contained one derivative chromosome 6. H9 and H9+Dox cells were all normal whereas one out of 15 LU07 cells and one out of 20 LU07+Dox cells displayed an additional chromosome 12, respectively (Figure S1). Long-term exposure with Dox did not lead per se to increased aneuploidies, since undifferentiated LU07+Dox cells maintained in vitro for more than 6 weeks with Dox were karyotypically normal (data not shown).
Teratoma Formation and Analysis
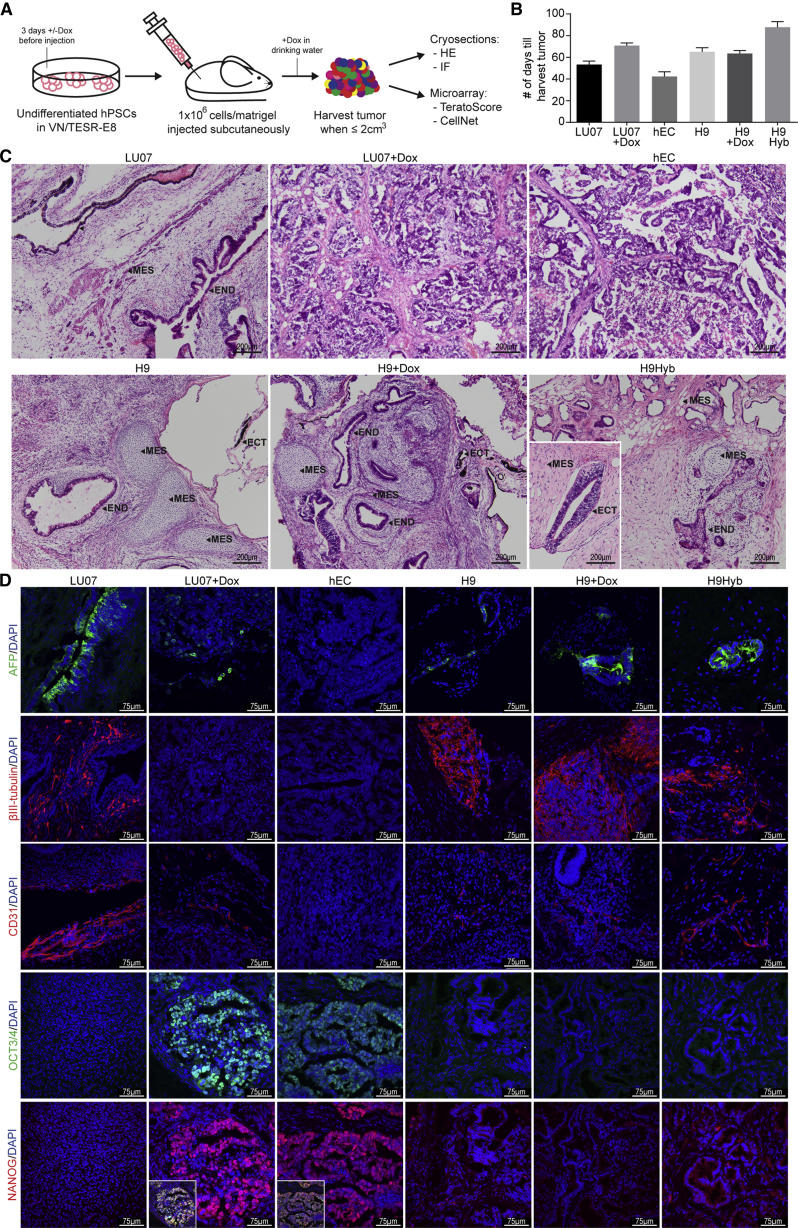
To test the differentiation capacity of hPSCs and hECs in the conventional in vivo Teratoma assay, we injected 1 million undifferentiated cells in the presence of Matrigel subcutaneously into the flank of immunodeficient mice. In initial experiments we found the NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) strain of mice to be more permissive for teratoma formation than NOD.CB17-Prkdcscid/J (NOD-SCID) (data not shown). When indicated, LU07 cells were pretreated with Dox for 3 days prior to injection, and mice received Dox in the drinking water 1 week before injection and during the whole period of tumor formation (LU07+Dox) (Figure 2A). To test whether Dox had any effects independent of transgene induction, we carried out similar experiments with H9 cells in the presence of Dox (H9+Dox). Xenografts were harvested between 31 and 112 days when reaching a maximum volume of 2 cm3. The administration of Dox did not significantly alter the growth rate of tumors (Figure 2B). For histological analysis, cryosections of one tumor per cell line were stained with H&E and examined by a certified pathologist.
Figure 2.
In Vivo Differentiation with the Teratoma Assay
(A) Schematic of the experimental procedure for teratoma induction.
(B) Days of xenograft growth until harvest (±SEM). For each cell line the same batch of cells were injected into six to eight mice (n = 7 except for H9Hyb, n = 8 and hEC, n = 6).
(C) Representative sections of H&E-stained xenografts. Arrowheads indicate derivatives of mesoderm (MES), ectoderm (ECT), and endoderm (END). Scale bars: 200 μm.
(D) IF staining using antibodies against βIII-tubulin (ectoderm), α-fetoprotein (AFP, endoderm), PECAM-1 (mesoderm), OCT3/4, and NANOG (undifferentiated cells). Insets: overlay of OCT3/4 and NANOG. Nuclei were stained with DAPI. Scale bars, 75 μm.
See also Figure S2.
H9, H9Hyb, and LU07 xenografts all contained differentiated structures representing the three germ layers (Figure 2C; neural rosettes and retinal pigmented epithelium [ectoderm], intestinal epithelium [endoderm], cartilage, bone, fat, and muscle [mesoderm]). The H9+Dox and the H9 teratomas had similar histological features. By contrast, LU07+Dox as well as the hEC tumor were principally composed of an embryonal carcinoma-like component, without any clearly differentiated tissues (Figure 2C). Accordingly, the hEC and LU07+Dox tumor were diagnosed as ”teratocarcinoma” (Damjanov and Andrews, 2007) or “embryonal carcinoma” according to the World Health Organization (WHO) guidelines (Williamson et al., 2017).
Since differentiated single cells or small groups are difficult to identify in H&E staining, we performed IF staining with antibodies directed against βIII-tubulin (ectoderm), human α-fetoprotein (endoderm), and human PECAM-1 (mesoderm). H9, H9+Dox, H9Hyb, and LU07 teratomas all contained areas with neurons, structures of endodermal origin, and endothelial cells (Figure 2D). By contrast, none of these cell types could be detected in the hEC tumor. In the LU07+Dox xenograft, βIII-tubulin-expressing cells were also undetectable. Endoderm and mesoderm were evident as a small number of scattered single cells (Figure 2D), indicating that their differentiation was impaired. To determine whether tumors still contained undifferentiated cells, we stained cryosections for pluripotency markers OCT3/4 and NANOG. In the hEC tumor, the great majority of cells co-expressed OCT3/4 and NANOG (Figures 2D and S2A). In the LU07+Dox xenograft, embryonal carcinoma-like cells expressed OCT3/4 and NANOG whereas these markers were absent in the surrounding stromal cells (Figures 2D and S2A). A significant proportion of the embryonal carcinoma-like cells also showed expression of the 2A peptide, indicating the activation of transgenes by Dox (Figure S2B). By contrast, OCT3/4 and NANOG were not detected in the sections of LU07, H9, H9+Dox, and H9Hyb teratomas. In summary, the reactivation of transgenes largely prevented differentiation in the LU07+Dox xenograft as determined by histological analysis. Similarly, the hEC tumor was completely undifferentiated and both tumors were classified as malignant. By contrast, the teratomas of LU07, H9, H9+Dox, and H9Hyb lines lacked undifferentiated cells and contained typical derivatives of the three germ layers.
Quantification of differentiated derivatives in teratomas by histological analysis is very laborious and subject to sampling error (Tsankov et al., 2015). This is due to the heterogeneous composition of the tumor (Figure 2C), difficulties in reliably determining cell identity in H&E staining, the analysis of only a limited number of markers by IF staining, and potential contamination of the xenograft by host mouse cells (e.g., endothelial cells of invading blood vessels).
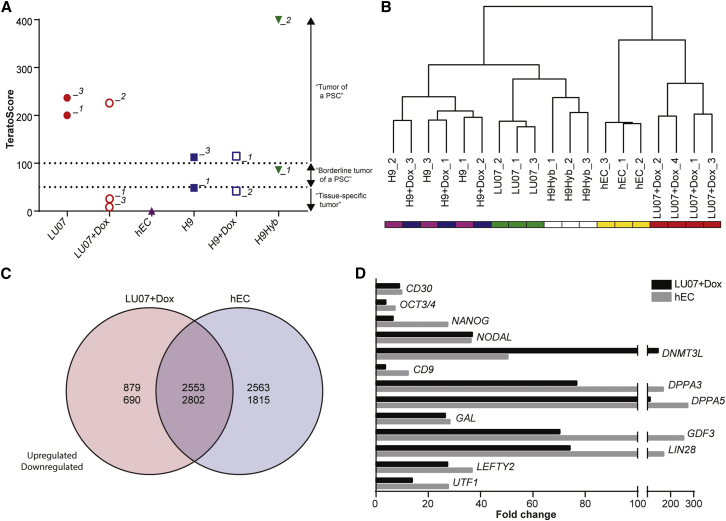
Recently an algorithm for the quantification of teratomas, called TeratoScore, was described (Avior et al., 2015). The TeratoScore is calculated based on the expression of a set of 100 genes representing ectoderm, endoderm, mesoderm, and extraembryonic tissue: scores higher than 100 indicate an hPSC-derived teratoma, whereas a value lower than 50 marks a tissue-specific tumor, e.g., a medulloblastoma. Values between 50 and 100 are considered borderline for hPSCs (Avior et al., 2015). RNA was extracted from whole tumors, and processed and analyzed with Affymetrix Human Genome (HG)-133 arrays. Both LU07 xenografts scored in the range of typical hPSC-derived teratomas whereas the TeratoScore results for teratomas derived from the other lines were more variable (Figure 3A). Surprisingly, half of the H9 and H9+Dox teratomas scored below the borderline (49 and 42, respectively). Evaluation of the individual TeratoScores showed that both xenografts mainly consisted of ectoderm and only little meso-, endo-, or extraembryonic tissue (Figure S3A). In general ectoderm was the most prevalent germ layer with the exception of H9Hyb_2 teratoma, which contained significant amounts of mesoderm and endoderm (Figure S3A). As expected, the tumor of the differentiation-defective hECs scored lowest (0.23). The scores for two LU07+Dox tumors were also low (LU07+Dox_1, 24; LU07+Dox_3, 9) whereas the score of the LU07+Dox_2 sample (226) was similar to that of LU07.
Figure 3.
Microarray Analysis of Teratomas
(A) TeratoScore results: >100, teratoma of a typical hPSC; 100–50, borderline hPSC teratoma; <50, tissue-specific tumor.
(B) Hierarchical clustering based on global gene expression for the same xenografts as in (A) with additional samples.
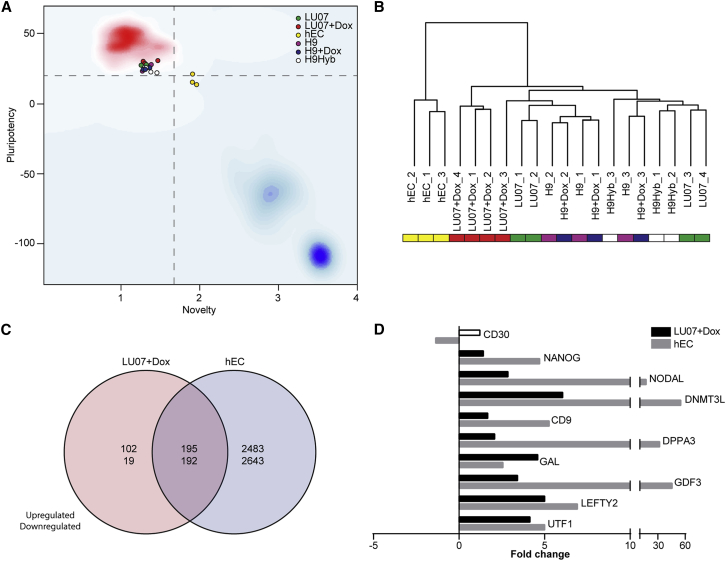
(C) Venn diagram of differentially expressed genes of hEC and LU07+Dox tumors compared with H9, H9+Dox, and LU07 teratomas (false discovery rate adjusted p value <0.05, log2 fold change >0.5).
(D) Significantly upregulated pluripotency- and/or cancer-associated genes in LU07+Dox and hEC tumors in fold change compared with H9, H9+Dox, and LU07 teratomas (FDR adjusted p < 0.05).
See also Figure S3.
Since the TeratoScore 100-gene list lacks markers of undifferentiated cells, we determined the expression of endogenous NANOG and the transgenes by qPCR in the xenografts. In the LU07_01 teratoma, NANOG levels were low (Figure S3B). By contrast, in the LU07+Dox_2 tumor, which had a similar TeratoScore, NANOG and transgenes were significantly higher, indicating that a fraction of the cells were still undifferentiated. The NANOG and transgene levels were more elevated in LU07+Dox_1 and _3 tumors (Figure S3B), which is in line with their low TeratoScores. In the H9 and H9+Dox tumors, which did not qualify as typical hPSC-derived teratomas either, NANOG levels were low (Figure S3C), indicating that the great majority of the cells had differentiated. Taken together, the TeratoScore algorithm confirmed our histology data showing the potential for three-germ-layer differentiation for LU07 and the nullipotency of hECs. TeratoScore results were variable for H9, H9+Dox, and H9Hyb xenografts but at least one tumor for each line qualified as typical teratoma. Only two out of three LU07+Dox tumors gave a low TeratoScore, which was possibly linked to variable levels of transgene induction. Importantly, the lack of pluripotency markers in the TeratoScore 100-gene list can lead to similar scores for xenografts, which grossly differ in the proportion of undifferentiated cells.
For each cell line we analyzed the tumors used for TeratoScore as well as additional samples with the more commonly used Illumina HT-12 platform. In line with our histology data, two clusters emerged based on whole transcriptome data: the differentiation-defective and malignant hEC and LU07+Dox tumors on the one hand and the teratomas derived from H9Hyb, H9, H9+Dox, and LU07 on the other (Figure 3B). Within each cluster, xenografts generated from the same cells were highly similar. Compared with normal teratomas (LU07, H9, H9+Dox), more than 6,500 genes were differentially expressed in the LU07+Dox or the hEC tumor and a large proportion were shared by both (Figure 3C). Various differentially expressed genes have known roles in pluripotency and malignancy, for example NANOG, OCT3/4, CD30, UTF1, and LIN28 (Figure 3D). Of note, the upregulated OCT3/4 in the LU07+Dox tumor is endogenous since the mouse transgenes are not detected by the human-specific microarray platform. The microarray data are in line with our IF staining results for OCT3/4 and NANOG (Figure 2D). In addition, immunohistochemistry staining showed increased NANOG and CD30 protein levels in the LU07+Dox and hEC xenografts (Figure S2A). It has been shown that CD30, a member of the tumor necrosis factor receptor family, is expressed on transformed hESCs (Herszfeld et al., 2006) and is a well-recognized diagnostic marker for hECs (Ulbright et al., 2014).
Since LU07+Dox and hEC xenografts showed histologically malignant features, we analyzed the tumors with a recently published qPCR assay for aneuploidies commonly found in hPSC cultures (Baker et al., 2016). As shown in Figure S2C, hEC tumors contained additional copies of (partial) chromosomes 1, 12, and 20. By contrast, aneuploidies were not detected in the LU07+Dox xenografts or in any of the other teratomas (Figure S2C).
Taken together, the microarray data confirmed that LU07+Dox tumors with reactivated transgenes and hEC tumors are highly undifferentiated and display features of malignancy. However, common aneuploidies were only identified in the hEC xenografts.
In Vitro Differentiation
Recently, the hPSC ScoreCard assay was proposed as a surrogate for the Teratoma assay to assess the functional pluripotency of hPSCs. It quantifies hPSC in vitro differentiation potential by measuring the expression of nine self-renewal genes and of 70 genes representing specific lineages by qPCR. The hPSC ScoreCard algorithm compares the expression levels with those of 13 undifferentiated standard hPSCs, including H9, then provides a single score for the query sample for self-renewal and each of the three germ layers (Bock et al., 2011, Tsankov et al., 2015).
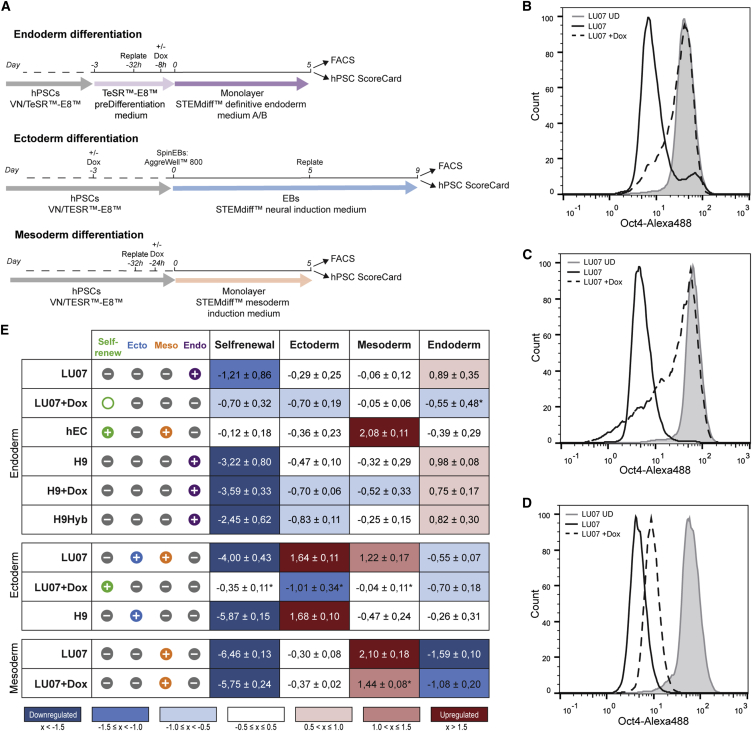
We first performed endodermal differentiation using a monolayer protocol with Dox added to H9 and LU07 where indicated (Figure 4A). After 5 days, cells were analyzed by FACS and the hPSC ScoreCard assay. As expected, no reduction of OCT3/4-expressing cells was observed in hECs (87% ± 3.8%, Figure S4A). LU07+Dox cells maintained a similar percentage of OCT3/4+ cells (85% ± 2.6%) indicating impaired differentiation (Figures 4B and S4A). By contrast, the proportion of OCT3/4+ cells was significantly smaller for LU07 cells (58% ± 8.1%) as well as for H9, H9+Dox, and H9Hyb (41% ± 15.2%, 48% ± 18.2%, and 22% ± 23.0%, respectively, Figure S4A). For transcriptional analysis, total RNA was isolated and qPCR performed with the commercially supplied hPSC ScoreCard plates. As shown in Figure 4E, LU07, H9, H9+Dox, and H9Hyb all downregulated self-renewal genes and exclusively differentiated into endoderm. All lines had similar endodermal scores despite the previously reported differentiation bias of H9Hyb (toward mesendoderm) and H9 cells (toward ectoderm), suggesting that perhaps newer endoderm differentiation protocols are more effective. By contrast, levels of self-renewal genes were unchanged or only slightly downregulated in hECs and LU07+Dox cells, respectively. Both cell lines were unable to give rise to endoderm (Figure 4E). Neither ectoderm nor mesoderm was induced in LU07+Dox. Interestingly, for hECs the mesoderm score was upregulated. We found a similar mesoderm score for hECs cultured in maintenance medium (data not shown). RGS4 and NKX2.5 were among the highly upregulated ScoreCard mesodermal genes (data not shown). This is in line with earlier findings indicating that RGS4 and NKX2.5 expression is more than 50-times higher in the same hEC line compared with a standard hESC line (Josephson et al., 2007). Thus, despite the expression of self-renewal genes, hECs may co-express certain mesodermal genes independent of the culture condition.
Figure 4.
hPSC ScoreCard Analysis of In Vitro Differentiations
(A) Schematics of endodermal, ectodermal, and mesodermal differentiation procedures.
(B–D) Representative FACS histogram of OCT3/4 in LU07 cells before differentiation (gray area) and in LU07 (black line) and LU07+Dox cells (dashed line) at the end of endodermal (B), ectodermal (C), and mesodermal (D) differentiation.
(E) hPSC ScoreCard results for endodermal differentiation (upper panel), ectodermal differentiation (middle panel), and mesodermal differentiation (lower panel). Left: hPSC ScoreCard result icons “+” (positive), “O” (borderline), or “–” (negative) are displayed and color coded green (self-renewal), blue (ectoderm), orange (mesoderm), and purple (endoderm). Icons represent the average of biological repeats (endoderm: n = 4 except for hEC, H9+Dox, and H9Hyb [n = 3]; ectoderm: LU07, n = 5; LU07+Dox, n = 3, H9, n = 2; mesoderm: n = 4). Right: Average scores (±SEM) of the same differentiations. Blue, downregulated; white, unchanged; red, upregulated. ∗p < 0.05.
See also Figure S4.
To test the capacity for ectodermal differentiation, we next performed monolayer differentiation (Figure S4B). However, hPSC ScoreCard analysis showed that H9Hyb was the only line with an elevated average score for ectoderm (Figure S4C) despite a reported mensendodermal differentiation bias (Qin et al., 2014). By contrast, H9+Dox cells did not give rise to ectoderm and for H9 and LU07 the average ectoderm score was low to borderline, respectively (Figure S4C). Previously, H9 embryoid bodies (EBs) have been shown to differentiate efficiently into the neuroectodermal lineage (Bock et al., 2011) suggesting that this particular ectodermal differentiation protocol was not optimal for analysis by the hPSC ScoreCard. As an alternative method, we used the Stemdiff Neural Induction system whereby spin-EBs are cultured in suspension for 5 days followed by 4 days of adherent culture (Figure 4A). FACS analysis revealed a significant reduction in OCT3/4+ cells for LU07, whereas in the presence of Dox the proportion of OCT3/4-expressing cells was similar to that of undifferentiated cells (42% ± 9.8% and 92.8% ± 2.3%, respectively; Figures 4C and S4A). In line with this, the self-renewal score for LU07 was low whereas self-renewal markers were maintained in LU07+Dox (Figure 4E). LU07+Dox did not give rise to ectoderm or derivatives of any other germ layer. By contrast, the differentiation of LU07 was confirmed by an elevated score for ectoderm as well as for mesoderm. The latter may be due to formation of neural crest cells in parallel with the neural progenitors. H9 cells differentiated exclusively into ectoderm (Figure 4E).
Finally, we tested the mesodermal differentiation capacity of LU07 and LU07+Dox using a monolayer differentiation protocol (Figure 4A). At day 5 of differentiation, the percentage of cells expressing high levels of OCT3/4 was reduced in both LU07 and LU07+Dox (6% ± 1.4% and 53% ± 9.2%, respectively; Figure S4A) compared with undifferentiated cells. Nevertheless, LU07+Dox cells still showed moderate OCT3/4 expression levels whereas the expression was low in LU07 cells (Figure 4D). The hPSC ScoreCard analysis indicated a pronounced downregulation of self-renewal genes in cells with and without Dox (Figure 4E). Although Dox was unable to prevent the differentiation of LU07+Dox cells toward mesoderm, the average mesodermal induction was significantly lower than in LU07 cells (Figure 4E). Taken together, hPSCs efficiently differentiated into endoderm (H9, H9+Dox, H9Hyb, LU07), ectoderm (H9, LU07), and mesoderm (LU07). By contrast, the reactivation of transgenes by addition of Dox maintained LU07+Dox in the undifferentiated state and prevented differentiation into endoderm or ectoderm. However, mesoderm induction was only slightly reduced. In conclusion, the ScoreCard assay can assess pluripotency at a functional level. By providing a score for self-renewal and each of the three germ layers, the hPSC ScoreCard assay can distinguish normal hPSCs from cells with a differentiation defect. Our in vitro differentiation data for LU07 and LU07+Dox cells confirmed the teratoma data with respect to pluripotency.
Pluripotency Analysis of Undifferentiated Cells
A microarray-based tool using undifferentiated hPSCs has also been described as another in vitro alternative to the Teratoma assay (Muller et al., 2011, Muller et al., 2012). This PluriTest algorithm uses genome-wide transcriptional profiles of more than 260 validated hPSCs (223 hESCs and 41 hiPSCs) as well as differentiated cell types and developing and adult tissues as a reference dataset for comparison with the query sample. A Pluripotency Score above 20 and a Novelty Score below 1.67 indicate that cells resemble typical hPSCs and that they are similar to normal hPSCs, respectively (Muller et al., 2011, Muller et al., 2012). Analysis of undifferentiated cells by the PluriTest algorithm revealed that H9, H9+Dox, H9Hyb, LU07, and LU07+Dox cells all resembled validated normal hPSCs (Figure 5A). Cells with or without Dox treatment (3 days) were indistinguishable and had similar Pluripotency and Novelty Scores (Table S1). By contrast, all hEC samples clustered separately and had a borderline or low Pluripotency Score and an elevated Novelty Score (Figure 5A and Table S1). This is in line with our own and published data showing that hECs have a highly aberrant karyotype (Figure S1A) and that their expression levels of pluripotency markers are markedly different from normal hPSCs (Josephson et al., 2007, Muller et al., 2011). On the other hand, the PluriTest algorithm did not indicate that the Novelty or Pluripotency Score was altered for the tetraploid H9Hyb cells (Figure 5A and Table S1). In fact the global gene expression pattern of hPSC hybrids has been shown to be highly similar to that of their diploid parental hPSCs (Qin et al., 2014).
Figure 5.
PluriTest and Microarray Analysis of Undifferentiated Cells
(A) PluriTest results (n = 4 for LU07 and LU07+Dox; n = 3 for hEC, H9, H9+Dox, H9Hyb, biological repeats). The background encodes an empirical density map indicating pluripotency (red) and novelty (blue); thresholds for pluripotency (20, horizontal) and novelty (1.67, vertical) are indicated with dashed lines.
(B) Hierarchical clustering based on global gene expression for the same samples as in (A).
(C) Venn diagram of up- and downregulated differentially expressed genes in hECs and LU07+Dox compared with undifferentiated H9, H9+Dox, and LU07 (filled bars: FDR adjusted p < 0.05, log2 fold change >0.5).
(D) Significantly up- and downregulated pluripotency- and/or cancer-associated genes in undifferentiated LU07+Dox cells and hECs in fold change compared with H9, H9+Dox, and LU07 cells (filled bars: FDR adjusted p < 0.05; empty bar, not significant).
See also Table S1.
Since the PluriTest algorithm gave comparable results for all hPSCs except the hECs, we asked whether whole transcriptome analysis would reveal subtle differences, especially for the LU07+Dox cells with the reactivated transgenes. Hierarchical clustering analysis of whole transcriptome data confirmed that hECs are most distinct from H9Hyb, H9, and LU07 with or without Dox treatment. However, within the second cluster, three out of four LU07+Dox samples represented a subgroup slightly different from non-treated LU07 and the other hPSC lines (Figure 5B). Compared with H9, H9+Dox, and LU07, we found more than 500 genes to be upregulated or downregulated in undifferentiated LU07+Dox cells, of which 387 overlapped with hECs, respectively (Figure 5C). Of note, the human-specific Illumina microarray used for PluriTest cannot detect the Dox-inducible transgenes, which are encoded by mouse cDNAs. The microarray expression levels of the endogenous versions of the reprogramming factors (OCT3/4, MYC, KLF4, SOX2) were not significantly different between LU07 and LU07+Dox (data not shown and Figure 1D). However, additional pluripotency-associated markers including endogenous NANOG, UTF1, GDF3, GAL, LEFTY2, CD9, and NODAL (International Stem Cell Initiative et al., 2007) were significantly upregulated in LU07+Dox cells (Figure 5D). The same genes (except GAL) were expressed at even higher levels in hECs. In hECs approximately 5,500 genes were up- or downregulated compared with H9, H9+Dox, and LU07 cells (Figure 5C), and it seems that this higher number of deregulated genes led to borderline or low Pluripotency Scores (Figure 5A and Table S1).
Taken together, the Pluripotency and the Novelty Score designated hECs as being abnormal. By contrast, PluriTest qualified H9, H9+Dox, H9Hyb, LU07, and LU07+Dox cells as pluripotent and normal based on their gene expression patterns in the undifferentiated state. However, the PluriTest was unable to reveal subtle differences in the expression of endogenous pluripotency-associated genes between LU07 and LU07+Dox, which together with the reactivated reprogramming factors are likely to affect functional pluripotency.
Discussion
In a side-by-side comparison we analyzed several hPSC lines with distinct differentiation capacities using the Teratoma assay and various in vitro alternatives such as the PluriTest and the hPSC ScoreCard. With the LU07+Dox cells we developed an experimental model of differentiation-defective hPSC lines (Figure 1), which we used to challenge current state-of-the-art pluripotency assays.
H9 hESCs, H9Hyb hPSCs, and LU07 hiPSCs all formed teratomas containing the three germ layers and lacking undifferentiated cells as shown by H&E and IF staining (Figure 2). By contrast, hEC and LU07+Dox tumors closely resembled each other in being mainly composed of embryonal carcinoma-like cells still expressing pluripotency markers.
Quantitative analysis of the xenografts by TeratoScore confirmed that LU07 hiPSCs, H9 hESCs, and H9Hyb hPSCs can give rise to typical hPSC-derived teratomas (Figure 3A). TeratoScore also confirmed the nullipotency of hECs and a differentiation defect in two out of three LU07+Dox tumors. The third LU07+Dox tumor received a TeratoScore similar to that of the LU07 teratoma. However, we found that these tumors varied substantially in their NANOG expression levels, indicating that a significant proportion of undifferentiated cells was still present in the LU07+Dox tumor. The fact that pluripotency markers are not included in the TeratoScore 100-gene list can lead to similar scores for fully and partially differentiated tumors. Moreover, we found variable TeratoScore results for H9, H9+Dox, and H9Hyb xenografts, indicating a typical teratoma, a borderline tumor, or even a primary tumor (Figure 3A). It seems advisable that at least two teratomas derived from the same cells should be analyzed with TeratoScore to reduce the risk of falsely categorizing cells as non-pluripotent.
In general, our histologically classical defined teratomas containing tissues from all three germ layers scored lower than the average TeratoScore reported by Avior et al. (2015). Although attempts have been made, the Teratoma assay has not been standardized to date (Gropp et al., 2012). The resulting variability in experimental procedures and histological interpretation has hampered the comparison of results between different laboratories (Muller et al., 2010). Indeed, Avior et al. (2015) cultured hPSCs on MEFs and injected 3 × 106 cells under the kidney capsule of NOD-SCID mice, whereas our tumors were induced with 1 × 106 TESR-E8 cells injected subcutaneously into NSG mice.
Our histological analysis of tumors revealed that the LU07+Dox tumor resembled the hEC tumor and was therefore classified as malignant teratoma or teratocarcinoma (Muller et al., 2010) or, according to the WHO nomenclature for human germ cell tumors, as a typical “embryonal carcinoma” (Williamson et al., 2017). We confirmed this diagnosis by immunohistochemistry staining for OCT3/4, NANOG, SOX2, anti-cytokeratin (AE1/AE3), and CD30 (Figure S2A and data not shown) (Ulbright et al., 2014). Characteristics of malignancy are rarely discussed when interpreting results from the Teratoma assay. We believe that the application of the WHO human germ cell tumor classification could be more useful for a detailed evaluation of elements that are associated with malignancy.
In addition to the histology data, clustering analysis of whole transcriptome data confirmed that all four LU07+Dox tumors were more similar to embryonal carcinoma-like tumors than to normal teratomas (Figure 3B). The transformation of LU07+Dox cells toward a malignant phenotype in vivo is likely to be caused by the prolonged reactivation of the reprogramming factors, which have all been shown to play a role in cancer. For example, c-MYC is a known oncogene and its abnormal expression has been shown to affect genomic integrity (Barlow et al., 2013). Indeed we identified several aneuploidies in hEC xenografts. However, LU07 or LU07+Dox tumors did not reveal any typical aneuploidies (Figure S2C) despite the fact that a fraction of the undifferentiated cells used for injection had an additional chromosome 12 (Figure S1). The qPCR assay used for detection of aneuploidies only covers (parts of) the chromosomes typically affected in hPSC cultures (chromosomes 1, 12, 17, and 20). Therefore, we cannot rule out that the malignant phenotype of LU07+Dox tumors is (partially) caused by karyotypic abnormalities affecting other regions or the remaining chromosomes.
In summary, the Teratoma assay can reveal the differentiation capacity of hPSCs in vivo and provides valuable information on the malignant potential of cells.
Our in vitro differentiation data, analyzed by the hPSC ScoreCard assay, confirmed the in vivo finding that LU07+Dox cells were unable to differentiate into endoderm and ectoderm and that they maintained the expression of pluripotency markers (Figure 4E). Furthermore, our hiPSC ScoreCard data confirmed that LU07 cells were able to differentiate into derivatives of all three germ layers in vitro (Figure 4E), which is in line with the teratoma data (Figure 2). However, the hPSC ScoreCard data did not confirm the differentiation biases reported by others previously or indicated by our TeratoScore data. For example, the H9 cells (with a reported differentiation bias toward ectoderm [Bock et al., 2011]) and LU07 cells (ectodermal differentiation bias indicated by TeratoScore) received endodermal scores similar to those of H9Hyb cells. Like TeratoScore, the hPSC ScoreCard is based on a limited number of genes representing the three germ layers. The question remains of whether assays which rely on a relatively restricted set of markers can indeed detect (subtle) lineage biases between cell lines.
It has been questioned whether in vitro assays that rely on complex and expensive protocols for directed differentiation or on spontaneous differentiations with EBs, which often contain undifferentiated cells in their core, would be useful for the assessment of pluripotency of hPSCs (Avior et al., 2015). Here we show that relatively simple, commercially available, short differentiation protocols based on cells in a monolayer (endoderm, mesoderm) or on short-term EBs consisting of a defined cell number (ectoderm) can be used for ScoreCard analysis. Each protocol required an initial optimization for each line in terms of cell number for plating or for incorporation into EBs, but then resulted in efficient differentiation toward the respective germ layer in multiple independent experiments.
Taken together, the hPSC ScoreCard data revealed the functional pluripotency of the LU07 cells and the differentiation deficiency of the LU07+Dox cells. The assay confirmed the teratoma data but is animal independent, much quicker, and quantitative.
An ideal assay to predict the functional pluripotency would be independent of in vitro and in vivo differentiation assays and exclusively rely on the analysis of undifferentiated cells. Currently there is no clear consensus on the minimal requirements for characterization of hPSCs at the molecular level (Chan et al., 2009). In addition, markers are demonstrated at mRNA or protein level and methods vary between qualitative (IF staining) or quantitative (FACS, qPCR). This lack of standardization makes it difficult to compare results between different research laboratories. In this respect the microarray-based bioinformatics assay PluriTest is a significant advance: it analyzes the global gene expression of a query sample and provides a quantitative result (Muller et al., 2011).
PluriTest has been shown to identify normal hPSC and highlight partially reprogrammed cells or nullipotent, karyotypically abnormal hECs as different from a large number of hPSC lines (Muller et al., 2011). However, LU07+Dox cells had a normal Pluripotency Score, despite their severely compromised differentiation capacities in vitro and in vivo. The inability of PluriTest to distinguish LU07+Dox cells with reactivated transgenes from LU07 cells partially results from the inducible reprogramming factors being encoded by mouse cDNAs, which are not detected by the human-specific microarray platform on which PluriTest is based. This could potentially concern any other integrating reprogramming vector with mouse transgenes.
A total of 508 endogenous genes were differentially expressed in LU07+Dox cells when compared with LU07, H9, and H9+Dox cells including several pluripotency- and/or cancer-related genes (Figure 5D). However, the fact that the Pluripotency Scores of LU07+Dox cells and LU07 cells were indistinguishable indicates that the PluriTest algorithm trained on a dataset from bona fide PSCs considered the global gene expression profile of LU07+Dox cells to be within the range of normal hPSCs. By contrast, in hECs, a total of more than 5,500 genes were different (Figure 5C). This higher degree of deregulation in global gene expression resulted in Pluripotency Scores at or below the threshold of normal hPSCs. Similar to LU07+Dox cells, a number of pluripotency-associated genes were upregulated in hECs; in general their induction was higher than in LU07+Dox cells (Figure 5D). It seems likely that the differentiation deficiency of LU07+Dox cells was caused by upregulation of the endogenous pluripotency-associated genes in combination with the reactivated transgenes. The overlap of approximately 400 deregulated genes shared between hEC and LU07+Dox cells suggest the possibility that given an externally validated dataset, additional models and scores within the PluriTest-framework could be trained to reliably detect other potentially differentiation-defective hPSC cell lines with properties similar to those of LU07+Dox cells.
In conclusion, the PluriTest provides quantitative information regarding whether a given cell line resembles normal hPSCs at a global molecular level. Higher-resolution RNA-sequencing data and the addition of epigenetic and microRNA profiles are likely to further improve the quality control of undifferentiated hPSCs. Short-term in vitro differentiation can reveal differences in functional pluripotency which are similar to the in vivo data. We therefore propose PluriTest in combination with the hPSC ScoreCard for routine characterization of hPSCs used for in vitro disease modeling and drug testing. Given the large numbers of hiPSC lines expected to be generated in the future, this would lead to a significant reduction of animal experiments and contribute to implementation of the “3Rs” policy (Replacement, Reduction, Refinement). Remarkably, the Teratoma assay is the only one of the three methods able to reveal malignant potential; this is a critical exclusion criterion for future hPSC clinical application. Indeed, the Teratoma assay has been proposed as a readout for tumorigenicity, although a quantitative analysis is currently lacking (Bulic-Jakus et al., 2016). However, the usefulness of a xenograft model for the prediction of malignancy in autologous clinical applications needs to be investigated much more in detail. In this respect mice with a humanized immune system may represent a further advance. Ideally, markers indicating potential malignancy could already be identified in undifferentiated cells.
In general, for better comparison of results between laboratories, all assays should be performed in standardized ways, e.g., with defined culture conditions for undifferentiated cells, a simple and robust set of in vitro differentiation protocols, and standardized procedures for teratoma induction and analysis.
Experimental Procedures
Full details are provided in Supplemental Experimental Procedures.
Cell Culture
All hPSCs were maintained on Vitronectin-XF in TESR-E8 medium (STEMCELL Technologies). LUMC007iCTRL01 was cultured in the absence of Dox unless otherwise stated. hECs were maintained in DMEM/F12 medium (Life Technologies) containing 10% FCS (Gibco).
In Vitro Differentiation Assays
Monolayer differentiation of hPSCs into neural stem cells was performed with Neural Induction Medium (Life Technologies). Neural progenitor cells were generated by using the STEMdiff Neural System EB protocol. For differentiation into endoderm, the TESR-E8 optimized STEMdiff Definitive Endoderm Kit was used. Mesoderm differentiation was performed using STEMdiff mesoderm induction medium (all from STEMCELL). All differentiations were performed according to the manufacturer's protocol. Dox was added when indicated (Figures 4A and S4B).
Teratoma Assay
Eight- to 10-week-old male NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Charles River) were used for injections. Cells (1 × 106) were injected subcutaneously in the flank region. Tumor growth was monitored weekly by palpation, and mice were euthanized when tumors reached a volume of ≤2 cm3. Animal experiments were approved by the Leiden University Medical Center (LUMC) Animal Ethics Committee. The LUMC is an institutional license holder according to Dutch Law on animal experimentation.
Immunofluorescent Staining and Immunohistochemistry
Staining procedures were performed according to standard procedures using 8-μm frozen sections or paraformaldehyde-fixed cultured cells. Primary antibodies were applied overnight at 4°C, followed by the secondary antibodies for 1 hr at room temperature. For antibodies, see Supplemental Experimental Procedures.
RNA Isolation and qRT-PCR
Xenografts were homogenized in RA1 buffer (Macherey-Nagel) with 1% β-mercaptoethanol using an Ultra-Turrax T8 homogenizer (IKA Labortechnik). An aliquot of the homogenate was used for RNA isolation using the NucleoSpin RNA kits (Macherey-Nagel), including a DNase-digestion step, according to the manufacturer's instructions. Quantitative expression analysis was performed on a Bio-Rad C1000 Thermal Cycler equipped with a CFX96/384 Real-Time System, with the iQ SYBR Green kit (Bio-Rad). Template cDNA was prepared from 1 mg of total RNA using the iScript cDNA synthesis kit (Bio-Rad). Expression of the target genes was normalized to hARP.
hPSC ScoreCard Assay
Template cDNA was prepared from 1 mg of RNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer's protocol. hPSC ScoreCard assays (Life Technologies) were run according to manufacturer's instructions on a Viia7 RT-PCR System (Applied Biosystems) using the ‘hpsc-ScoreCard-template-viia-7-384-well’ template.
Microarray Analysis
cRNA was labeled and hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays (Dutch Genomics Service & Support Provider, University of Amsterdam). RNA for analysis with PluriTest and from xenograft samples was labeled and hybridized onto the Illumina Human HT-12 v4 array by GenomeScan.
Author Contributions
M.J.B. performed experiments, analyzed data, and made figures. M.v.I. reviewed the microarray data analysis. B.J. performed microarray experiments. D.C.F.S. conceived the work, designed and performed experiments, analyzed and interpreted data, and participated in writing the manuscript. C.L.M. provided advice and contributed intellectually. C.F. designed and performed experiments, analyzed data, and wrote the manuscript. D.C.F.S. and C.F. contributed equally to this work.
Acknowledgments
We thank Dr. M. Zenke (RWTH Aachen University) and Dr. P.W. Andrews (University of Sheffield) for providing H9Hyb and 2102Ep hECs, respectively, and Dr. C. Dambrot for assistance in generating LUMC007iCTRL01 cells. We are grateful to Dr. I. Damjanov (Kansas University) and Dr. J.W. Oosterhuis (Erasmus Medical Center) for help with histology data analysis. We thank Dr. X. Yu (Amsterdam Medical Center) for his help with experiments in the initial phase of the project. S. Maas and S. Silvestri (LUMC) are thanked for preparation of cryosections, and J.A. Stoop (Erasmus Medical Center) for performing the CD30 immunohistochemistry staining. We are also grateful to Dr. K. Szuhai and D. de Jong for karyotype analysis and to Dr. R. Tsonaka for support with statistics (all LUMC). Special thanks to the LUMC hiPSC core facility staff for support. R. van der Straaten and J.J.M. Drabbels (LUMC) are thanked for providing access to the Viia7 instrument. Dr. Jan-Bas Prins (LUMC) is thanked for support and critical review of the animal experiments, and Dr. F.J. Müller (University Hospital Kiel) for critical reading of the manuscript. This work is part of the research program “Meer Kennis met Minder Dieren” (More Knowledge with Fewer Animals) with project number 40-42600-98-028, which is financed by the Netherlands Organization for Scientific Research (NWO).
Published: May 9, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.03.009.
Contributor Information
Daniela C.F. Salvatori, Email: d.c.f.salvatori@lumc.nl.
Christian Freund, Email: c.m.a.h.freund@lumc.nl.
Accession Numbers
The gene expression data reported in this work has been deposited at the GEO repository with accession number GEO: GSE95286.
Supplemental Information
References
- Anguera M.C., Sadreyev R., Zhang Z., Szanto A., Payer B., Sheridan S.D., Kwok S., Haggarty S.J., Sur M., Alvarez J. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avior Y., Biancotti J.C., Benvenisty N. TeratoScore: assessing the differentiation potential of human pluripotent stem cells by quantitative expression analysis of teratomas. Stem Cell Rep. 2015;4:967–974. doi: 10.1016/j.stemcr.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D., Hirst A.J., Gokhale P.J., Juarez M.A., Williams S., Wheeler M., Bean K., Allison T.F., Moore H.D., Andrews P.W. Detecting genetic mosaicism in cultures of human pluripotent stem cells. Stem Cell Rep. 2016;7:998–1012. doi: 10.1016/j.stemcr.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.H., Faryabi R.B., Callen E., Wong N., Malhowski A., Chen H.T., Gutierrez-Cruz G., Sun H.W., McKinnon P., Wright G. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Markoulaki S., van Wietmarschen N., Hoke H., Wu T., Ganz K., Akhtar-Zaidi B., He Y., Abraham B.J., Porubsky D. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulic-Jakus F., Katusic Bojanac A., Juric-Lekic G., Vlahovic M., Sincic N. Teratoma: from spontaneous tumors to the pluripotency/malignancy assay. Wiley Interdiscip. Rev. Dev. Biol. 2016;5:186–209. doi: 10.1002/wdev.219. [DOI] [PubMed] [Google Scholar]
- Buta C., David R., Dressel R., Emgard M., Fuchs C., Gross U., Healy L., Hescheler J., Kolar R., Martin U. Reconsidering pluripotency tests: do we still need teratoma assays? Stem Cell Res. 2013;11:552–562. doi: 10.1016/j.scr.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M., Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J.H., Faddah D.A., Buganim Y., Kim J., Ganz K., Steine E.J., Cassady J.P., Creyghton M.P. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Chan E.M., Ratanasirintrawoot S., Park I.H., Manos P.D., Loh Y.H., Huo H., Miller J.D., Hartung O., Rho J., Ince T.A. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Choi J., Lee S., Mallard W., Clement K., Tagliazucchi G.M., Lim H., Choi I.Y., Ferrari F., Tsankov A.M., Pop R. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley G.Q., Lensch M.W., Jaenisch R., Meissner A., Plath K., Yamanaka S. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell. 2009;4:200–201. doi: 10.1016/j.stem.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Damjanov I., Andrews P.W. The terminology of teratocarcinomas and teratomas. Nat. Biotechnol. 2007;25:1212. doi: 10.1038/nbt1107-1212a. [DOI] [PubMed] [Google Scholar]
- Dolgin E. Putting stem cells to the test. Nat. Med. 2010;16:1354–1357. doi: 10.1038/nm1210-1354. [DOI] [PubMed] [Google Scholar]
- Gertow K., Przyborski S., Loring J.F., Auerbach J.M., Epifano O., Otonkoski T., Damjanov I., Ahrlund-Richter L. Isolation of human embryonic stem cell-derived teratomas for the assessment of pluripotency. Curr. Protoc. Stem Cell Biol. 2007;Chapter 1 doi: 10.1002/9780470151808.sc01b04s3. Unit1B.4. [DOI] [PubMed] [Google Scholar]
- Gropp M., Shilo V., Vainer G., Gov M., Gil Y., Khaner H., Matzrafi L., Idelson M., Kopolovic J., Zak N.B. Standardization of the teratoma assay for analysis of pluripotency of human ES cells and biosafety of their differentiated progeny. PLoS One. 2012;7:e45532. doi: 10.1371/journal.pone.0045532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herszfeld D., Wolvetang E., Langton-Bunker E., Chung T.L., Filipczyk A.A., Houssami S., Jamshidi P., Koh K., Laslett A.L., Michalska A. CD30 is a survival factor and a biomarker for transformed human pluripotent stem cells. Nat. Biotechnol. 2006;24:351–357. doi: 10.1038/nbt1197. [DOI] [PubMed] [Google Scholar]
- Inoue H., Nagata N., Kurokawa H., Yamanaka S. iPS cells: a game changer for future medicine. EMBO J. 2014;33:409–417. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Stem Cell Banking Initiative Consensus guidance for banking and supply of human embryonic stem cell lines for research purposes. Stem Cell Rev. 2009;5:301–314. doi: 10.1007/s12015-009-9085-x. [DOI] [PubMed] [Google Scholar]
- International Stem Cell Initiative, Adewumi O., Aflatoonian B., Ahrlund-Richter L., Amit M., Andrews P.W., Beighton G., Bello P.A., Benvenisty N., Berry L.S. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Josephson R., Ording C.J., Liu Y., Shin S., Lakshmipathy U., Toumadje A., Love B., Chesnut J.D., Andrews P.W., Rao M.S. Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells. 2007;25:437–446. doi: 10.1634/stemcells.2006-0236. [DOI] [PubMed] [Google Scholar]
- Kyttala A., Moraghebi R., Valensisi C., Kettunen J., Andrus C., Pasumarthy K.K., Nakanishi M., Nishimura K., Ohtaka M., Weltner J. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Rep. 2016;6:200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Muller F.J., Goldmann J., Loser P., Loring J.F. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell. 2010;6:412–414. doi: 10.1016/j.stem.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Muller F.J., Schuldt B.M., Williams R., Mason D., Altun G., Papapetrou E.P., Danner S., Goldmann J.E., Herbst A., Schmidt N.O. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F.J., Brandl B., Loring J.F. StemBook. Harvard Stem Cell Institute; 2012. Assessment of Human Pluripotent Stem Cells with PluriTest. [PubMed] [Google Scholar]
- Ohnuki M., Tanabe K., Sutou K., Teramoto I., Sawamura Y., Narita M., Nakamura M., Tokunaga Y., Nakamura M., Watanabe A. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl. Acad. Sci. USA. 2014;111:12426–12431. doi: 10.1073/pnas.1413299111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passier R., Orlova V., Mummery C. Complex tissue and disease modeling using hiPSCs. Cell Stem Cell. 2016;18:309–321. doi: 10.1016/j.stem.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Qin J., Sontag S., Lin Q., Mitzka S., Leisten I., Schneider R.K., Wang X., Jauch A., Peitz M., Brustle O. Cell fusion enhances mesendodermal differentiation of human induced pluripotent stem cells. Stem Cells Dev. 2014;23:2875–2882. doi: 10.1089/scd.2014.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuhai K., Tanke H.J. COBRA: combined binary ratio labeling of nucleic-acid probes for multi-color fluorescence in situ hybridization karyotyping. Nat. Protoc. 2006;1:264–275. doi: 10.1038/nprot.2006.41. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tsankov A.M., Akopian V., Pop R., Chetty S., Gifford C.A., Daheron L., Tsankova N.M., Meissner A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 2015;33:1182–1192. doi: 10.1038/nbt.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbright T.M., Tickoo S.K., Berney D.M., Srigley J.R., Members of the ISUP Immunohistochemistry in Diagnostic Urologic Pathology Group Best practices recommendations in the application of immunohistochemistry in testicular tumors: report from the International Society of Urological Pathology consensus conference. Am. J. Surg. Pathol. 2014;38:e50–59. doi: 10.1097/PAS.0000000000000233. [DOI] [PubMed] [Google Scholar]
- Williamson S.R., Delahunt B., Magi-Galluzzi C., Algaba F., Egevad L., Ulbright T.M., Tickoo S.K., Srigley J.R., Epstein J.I., Berney D.M. The World Health Organization 2016 classification of testicular germ cell tumours: a review and update from the International Society of Urological Pathology Testis Consultation Panel. Histopathology. 2017;70:335–346. doi: 10.1111/his.13102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.