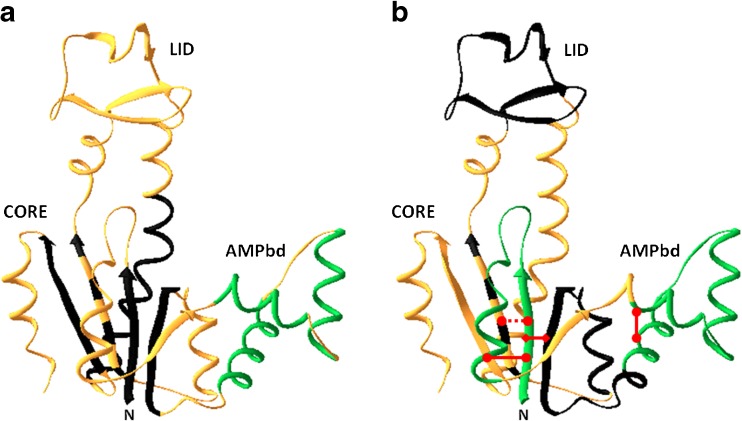
Fig. 3.
Ribbon diagram representing the fast and slow folding sub-domain loop and secondary structure elements marked on the native state chain fold of the backbone of the Escherichia coli AK molecule (PDB ID code: 4AKE). a Fast and slow folding secondary structure elements that were studied so far are color coded. Those that remain disordered at 5 ms after initiation of mixing into folding buffer (the dead-time) are colored in black. Those that gain native-like mean end-to-end distance at the 5 ms detection time are colored green. The three short helices in the AMPbind loop gain native-like end-to-end distance within the mixing dead-time (green). Parts of the chain that were not tested are colored in gold. Most of the segments which form secondary structures that constitute the core domain of AK are still disordered in the initial phase of the folding transition. b The non-local interactions between chain segments that are effective at the 5 ms collapsed ensemble are marked by a red line. At least three long loops are closed within the mixing time, i.e., at least within the initial 5 ms of refolding: the N terminal loop and the AMPbind loop (green), as well as a long loop between residues 1–75 (red diamond) that includes the two elementary loops. Chain segments that form a loop structure in the folded state but not in the intermediate state are colored in black