Figure 3.
PI3K/AKT/mTOR Activity Regulates Passive DNA Demethylation during VPA-Induced Differentiation
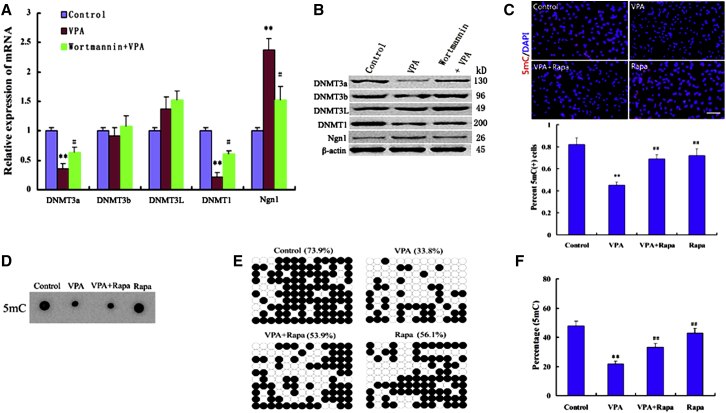
(A and B) Relative levels of mRNA and protein for different Dnmts and Ngn1 in NSCs treated with either VPA or VPA plus wortmannin. Results were normalized with β-actin. ∗∗p = 0.006, 0.004 < 0.01 compared with control; #p = 0.04, 0.03 < 0.05 compared with VPA. Data are from three independent experiments.
(C) Immunofluorescence staining for 5-mC showed that the intensity of VPA on 5-mC-immunostaining was attenuated by rapamycin (Rapa). Scale bar, 250 μm. ∗∗p = 0.002 < 0.01 compared with control; ##p = 0.006, 0.004 < 0.01 compared with VPA. Data are from three independent experiments.
(D) Dot blot analysis of 5-mC quantity during differentiation of NSCs. Dot blot performed by biotinylation of 5-mC followed by probing with Avidin-HRP. Dot blot representative of three independent experiments.
(E) Bisulfite DNA sequencing analysis to part of the Ngn1 promotor region in NSCs treated with VPA or rapamycin. Filled and open circles represent methylated and unmethylated CpGs, respectively. The percentage of total methylated CpGs for each analyzed region is given on top of each dataset.
(F) Relative quantitation of 5-mC within a specific locus was performed using the EpiMark 5-mC Analysis Kit followed by real-time PCR analysis. ∗∗p = 0.003 < 0.01 compared with control; ##p = 0.008, 0.006 < 0.01 compared with VPA. One-way ANOVA and Student's t test were used to determine the statistical significance. Data are from three independent experiments.