Fig. 3.
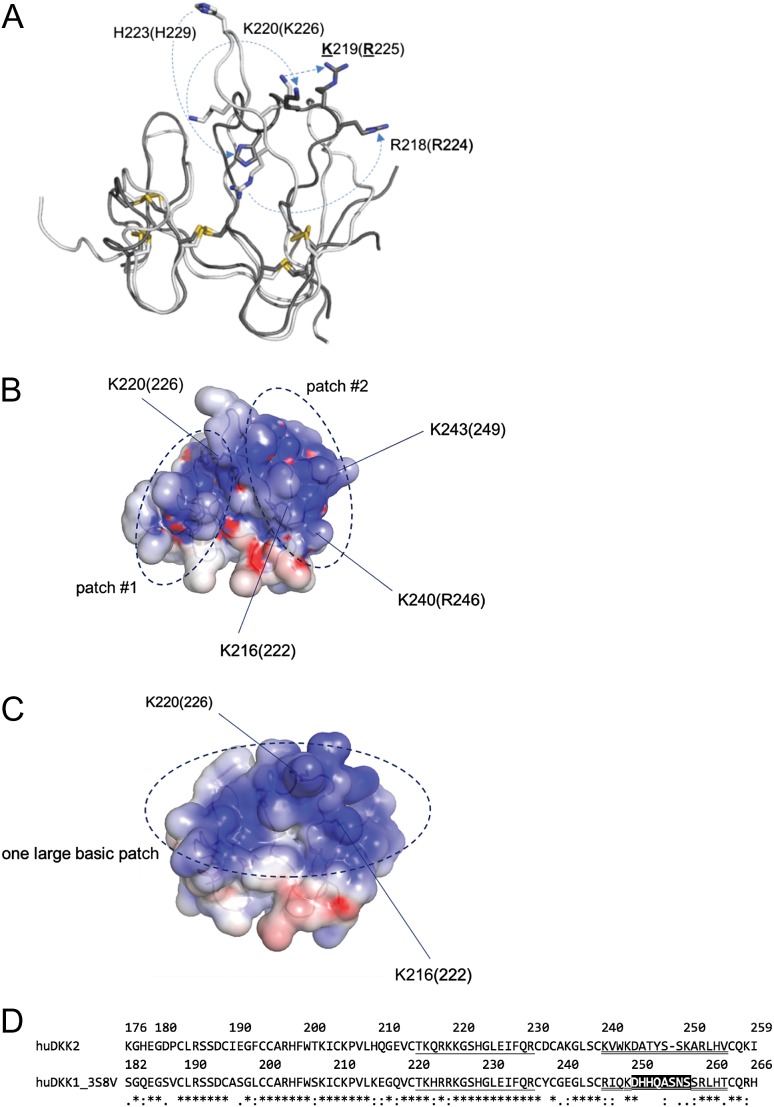
Rational design of HS-binding mutations in DKK2C2. (A) Schematic diagram showing conformational shifts between DKK2C2 (2JTK.pdb, white), based on an NMR structure at pH 5, and DKK1C2 (3S8V.pdb, dark gray), based on an X-ray structure at pH 8.8. Residue numbers are for DKK2C2 (open) and DKK1C2 (in parentheses). A number of basic residues undergo large conformational shifts between the two structures, such as H223 (229), K220 (226), and R218 (224). (B) Schematic representation showing the locations of basic patches #1 and #2 on the surface of DKK2C2 (2JTK.pdb). (C) Schematic representation showing the location of the large basic patch on the surface of DKK1C2 (3S8V.pdb) (all images in A–C were created in Pymol [The PyMOL Molecular Graphics System, Version 1.7.4.0 Schrödinger, LLC], electrostatic surfaces were created using APBS (Baker et al., 2001)). (D) Alignment of human DKK2C2 (as numbered in 2JTK.pdb) and DKK1C2 (as numbered in 3S8V.pdb); both 3S8V and a nearly identical structure of DKK1C2 bound to LRP6 (3S2K.pdb) have missing coordinates for residues D250–S257, which are indicated through a black background; the loop between conserved disulfide-bound cysteines C214 (220) and C231 (237), which undergoes the largest conformational shift between DKK2C2 and DKK1C2–LRP6 structures is single underlined; the C-terminal loop between conserved disulfide-bound cysteines C239 (245) and C256 (263) is double-underlined.