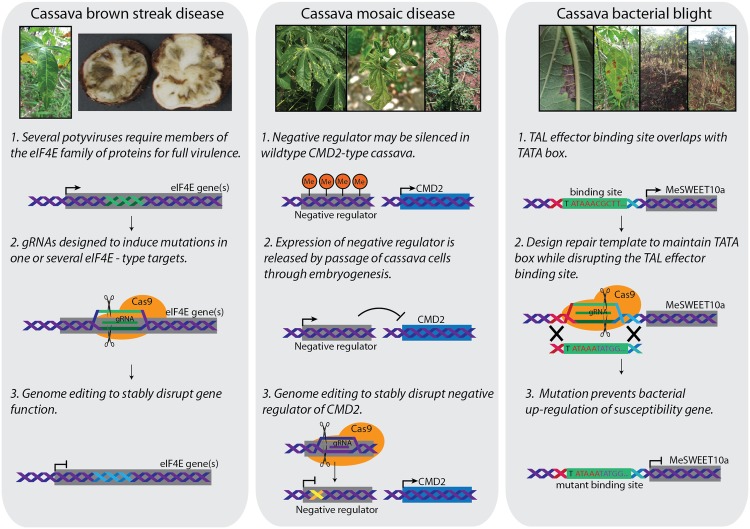
Fig 1. Genome-editing strategies to create resistance against three major pathogens of cassava.
(Left) Cassava brown streak disease (CBSD) is caused by two potyviruses. In other species, mutation of eukaryotic translation initiation factor 4E (eIF4E) family of genes promotes disease resistance. It is predicted that a similar strategy may be effective in cassava. (Middle) Cassava mosaic disease (CMD) is widely controlled by the yet to be identified CMD2 resistance mechanism. For unknown reasons, passage of cassava cells through somatic embryogenesis disrupts CMD2-mediated resistance. A potential explanation involves loss of methylation (Me) that releases expression of a negative regulator of CMD2. In this case, it should be possible to use guide RNAs complexed with Cas9 to introduce genetic changes that stably disrupt expression of the negative regulator. (Right) Cassava bacterial blight disease is caused by Xanthomonas axonopodis pv. manihotis (Xam). Xam injects transcription activator like (TAL) effectors into plant cells, which activate expression of MeSWEET10a by directly binding to its promoter. The binding site overlaps with the TATA box. Using homolog recombination, it would be possible to mutate the binding site, maintain the TATA box, and disrupt TAL effector binding.