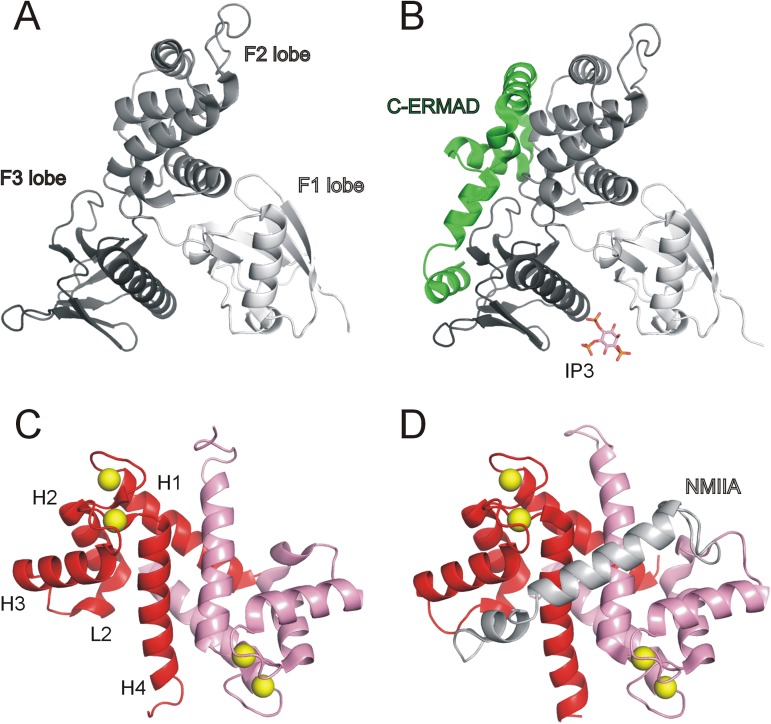
Fig 1. Structural overview of ezrin and S100A4 proteins.
(A) Crystal structure of the human ezrin N-ERMAD (PDB ID: 4RMA). (B) Crystal structure of full-length human ezrin (PDB ID: 4RM8). Note that the 160 residue-long α-helical domain, which connects the N-ERMAD and the C-ERMAD, and the N-terminal 39 residues of the C-ERMAD (Val477-Glu515) are not visible in the crystal structure. Crystal structure of mouse radixin N-ERMAD in complex with inositol-(1,4,5)-trisphosphate (IP3; PDB ID: 1GC6) was used to demonstrate the putative lipid-binding site of ezrin. (C) Crystal structure of calcium-bound S100A4 (PDB ID: 3C1V) and (D) calcium-bound S100A4 complexed with non-muscle myosin IIA (NMIIA) C-terminal fragment (PDB ID: 3ZWH).