Figure 1. Central Carbon Metabolites Tune Tobramycin Susceptibility in P. aeruginosa.
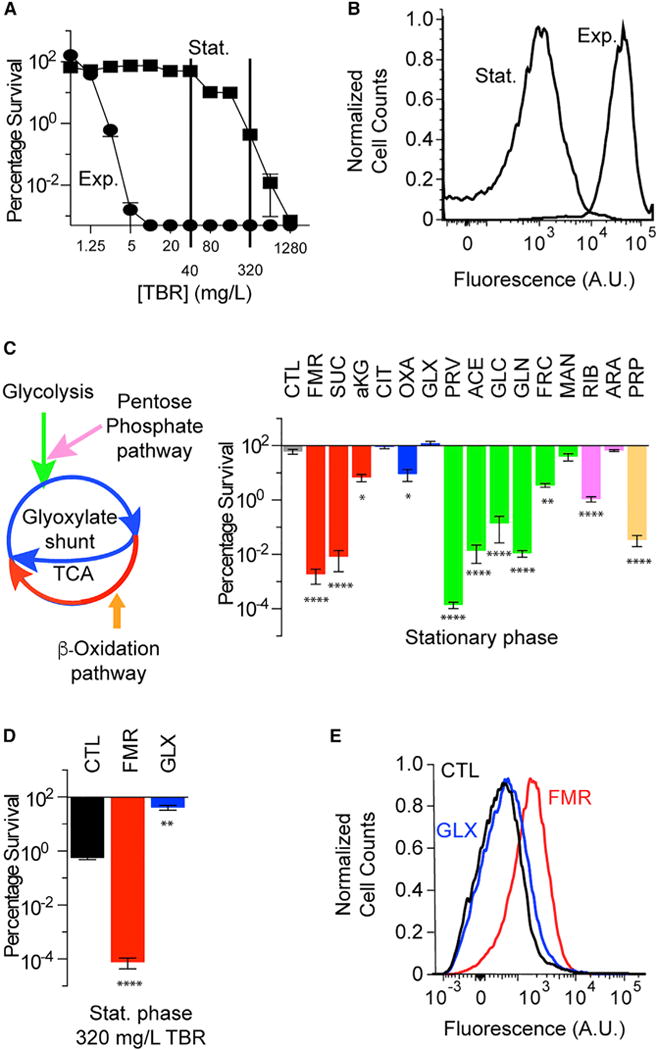
(A) Survival of exponential- (Exp.) and stationary-phase (Stat.) PAO1 cells following 4 hr tobramycin (TBR) treatment. Metabolically dormant stationary-phase cells exhibit phenotypic tolerance (n = 2).
(B) Representative flow cytometry quantification of exponential- and stationary-phase cells treated with 40 mg/L tobramycin-Texas red for 15 min. Cell counts are normalized to mode.
(C) Survival of stationary-phase cells following 4 hr treatment with 40 mg/L tobramycin and supplemented with various central carbon metabolites at a total carbon concentration of 60 mM. Supplemented metabolites are as follows: no carbon source (CTL), fumarate (FMR), succinate (SUC), α-ketoglutarate (aKG), citrate (CIT), oxaloacetate (OXA), glyoxylate (GLX), pyruvate (PRV), acetate (ACE), glucose (GLC), gluconate (GLN), fructose (FRC), mannitol (MAN), ribose (RIB), arabinose (ARA), or propionate (PRP). Cells are normalized to the pre-treatment CFU/mL.
(D) Survival of stationary-phase cells following 4 hr treatment with 320 mg/L tobramycin and supplementation with 15 mM fumarate or 30 mM glyoxylate or no supplement (control).
(E) Representative flow cytometry quantification of stationary-phase cells following 15 min treatment with 40 mg/L tobramycin-Texas red and supplementation with 15 mM fumarate or 30 mM glyoxylate or no supplement (control). Cell counts are normalized to mode. Statistical analysis of geometric means is depicted in Figure S1D.
Values in all bar graphs depict the mean ± SEM with significance reported as FDR-corrected p values in comparison with untreated control (CTL): *p ≤0.01, **p ≤ 0.001, ****p ≤ 0.0001 (n = 3).
