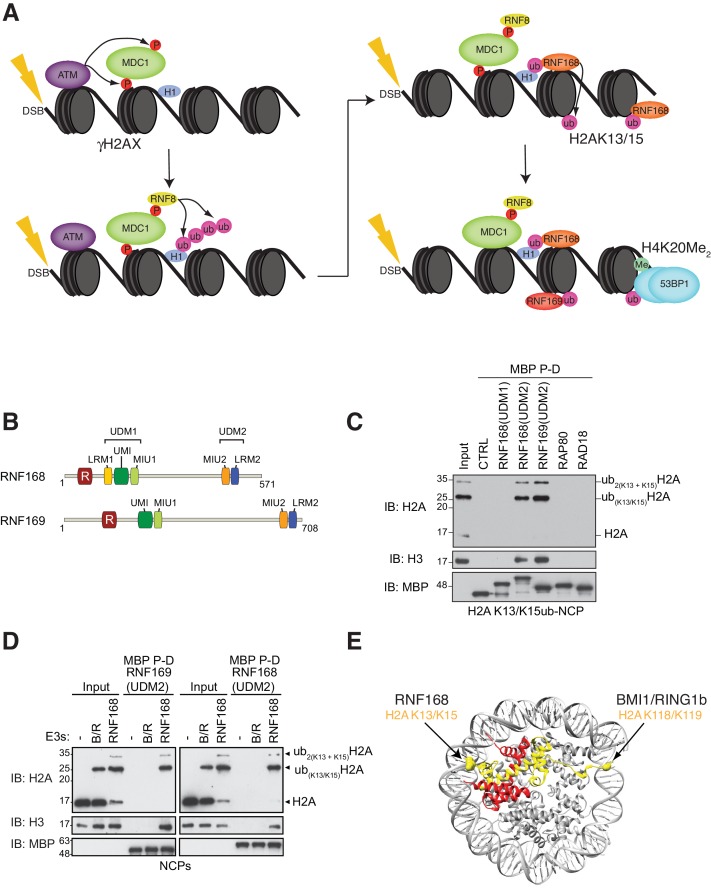
Figure 1. RNF168 and RNF169 bind RNF168-ubiquitylated NCPs.
(A) Schematic of RNF8-mediated DNA DSB repair pathway. ATM: Ataxia telangiectasia mutated, MDC1: mediator of DNA damage checkpoint 1, BRCT: breast cancer 1 C-terminal, H1: linker histone H1, RNF: ring finger proteins, 53BP1: p53 binding protein 1, Ub: ubiquitin, P: phosphate group, Me: methyl group. (B) Domain architecture of RNF168(1-571) and RNF169(1-708). Domains and motifs are indicated. R: RING domain, MIU: motif interacting with ubiquitin, UIM: ubiquitin-interacting motif, UMI: UIM-, MIU-related ubiquitin binding motif, LRM: LR motif. (C) MBP pull-down assays of RNF168-ubiquitylated nucleosome core particles (H2AK13/K15ub-NCP) with the indicated MBP fusion proteins (RNF168-UDM1(110–201), RNF168-UDM2(374–571), RNF169-UDM2(662–708), RAP80(60-124) and RAD18(201-240)). Input: 5% of the amount of ubiquitylated NCPs used in the pull-down. The migration of molecular mass markers (kDa) is indicated on the left. (D) Pull-down assays of NCPs ubiquitylated with the indicated E3s by either MBP–RNF169(UDM2) (left) or MBP–RNF168(UDM2) (right). A reaction without E3 (-) acts as a negative control. B/R: BMI1/RING1b. (E) Structure of the nucleosome (PDB: 2PYO [Clapier et al., 2008b]). One copy of H2A and H2B is labeled in yellow and in red, respectively. Lysines that are ubiquitylated by RNF168 (H2A K13/K15) and BMI1/RING1B (H2A K118/K119) are indicated in space filling representation.