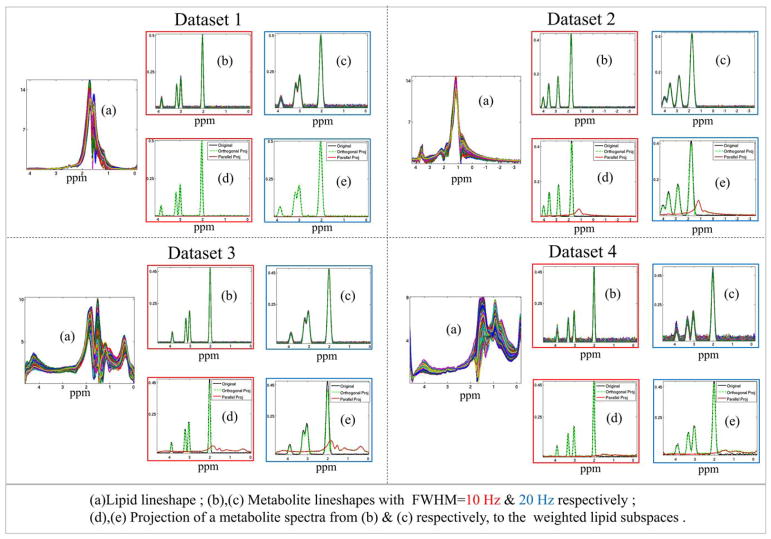
FIG. 4.
Spectral orthogonality of lipid and metabolite line shapes: Orthogonality between lipids and metabolites are demonstrated using four datasets. Dataset 1 and 2 are simulated using real lipid unsuppressed data of (40) and (17), which are distributed with the software. Dataset 3 and 4 are simulated using real data acquired from subject 1 and 2, respectively, without any lipid suppression. For each dataset, lipid line shapes generated from real data are shown in (a). The idealized metabolite spectra consisting of NAA, Creatine, and Choline peaks are simulated with FWHM=10 and 20 Hz (color-coded outline in red and blue, respectively) in (b) and (c), respectively. The metabolites are projected to a rank=20 weighted lipid subspace as given by Equation [14]. The parallel (in red) and orthogonal (in green) projection of a randomly selected metabolite spectra (in black) of the dataset, to the lipid subspace for the two simulations are plotted in (d) and (e), respectively. (For quantitative report of orthogonal projection energy, please refer to Supporting Fig. S1).