Figure 6.
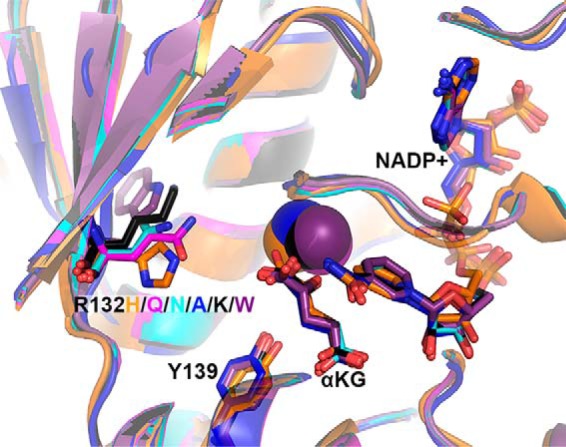
Structural models of experimental IDH1 mutants. The structure of R132H IDH1 complexed with αKG, NADP+, and Ca2+ (PDB 4KZO (27)) was used to model mutations of the tool IDH1 mutations. R132H IDH1 is shown in orange, R132Q in magenta, R132N in cyan, R132A in dark blue, R132K in black, and R132W in purple. Substrates and residues that are mutated are highlighted in stick format, as well as catalytic residue Tyr-139. Ca2+ is shown as a sphere. Ligand restraint generation and optimization of provided cif files were generated using eLBOW in the Phenix software suite (35), and mutations were made using Coot (54). Geometry Minimization (Phenix software suite) (35) was used to regularize geometries of the models, with 500 iterations and 5 macro cycles.
