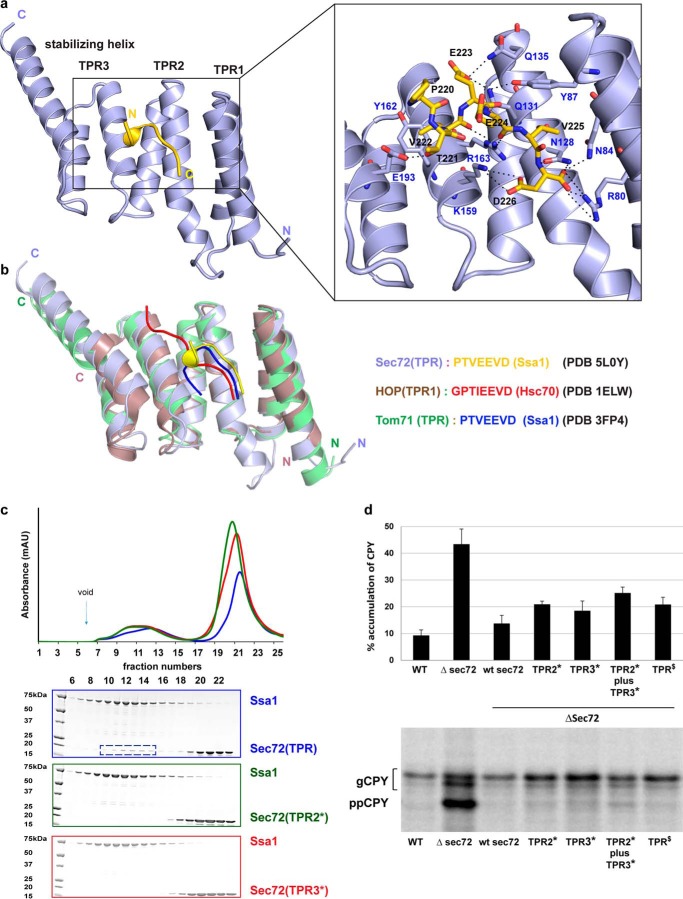
Figure 3.
Interaction of the TPR domain of Sec72 with the C terminus of Ssa1. a, crystal structure of a fusion of the TPR domain of Sec72 with a C-terminal Ssa1 peptide, both derived from Chaetomium thermophilum. Shown are ribbon diagrams with Sec72 in light blue and the peptide in yellow. The right panel gives a magnified view with the amino acids of the peptide and interacting TPR residues shown as sticks. Dashed lines indicate salt bridges and hydrogen bonds. b, structures of other TPR domains with bound C-terminal Hsp70 peptides. Note that similar peptides adopt different conformations when bound to different TPR domains. c, purified full-length Ssa1 was mixed with purified wild-type TPR domain of Sec72 and subjected to gel filtration. The absorbance at 280 nm was followed (blue curve in upper panel). Additional experiments were performed with TPR domains that carried mutations in either TPR2 (mutations of Asn-128, Gln-131; mutant TPR2*; green curve) or TPR3 and the stabilizing helix (mutation of Lys-159, Arg-163, and Asp-193; mutant TPR3*; red curve). For each experiment, fractions were analyzed by SDS-PAGE and EZBlue staining (lower panels; the colors correspond to the curves in the upper panel). The box indicates fractions containing the TPR domain of Sec72 co-eluting with Ssa1. d, the translocation of the post-translational substrate carboxypeptidase Y (CPY) was tested by incubating wild-type yeast cells or cells carrying Sec72 mutations with Trans35S-label. After immunoprecipitation, the samples were subjected to SDS-PAGE and autoradiography. The percentage of non-translocated CPY (prepro-CPY (ppCPY)) with respect to total CPY (glycosylated CPY (gCPY) plus ppCPY) was determined by densitometry from two (TPR2*) or four to six (all others) experiments, a representative of which is shown in the lower panel. The bars show mean (TPR2) and standard deviation (all others). TPR2* contains the mutations S111A and D114A in full-length S. cerevisiae Sec72 (equivalent to the TPR2* mutations in C. thermophilum). TPR3* contains the mutations R145A, D141A, and K175A (equivalent to the TPR3* mutations in C. thermophilum), and TPR$ contains D172R and E173K, equivalent to the E190R and E191R mutations in C. thermophilum (Fig. 4).