Abstract
Purpose
The purpose of this study was to identify mutations that cause non-syndromic male infertility using whole exome sequencing of family cases.
Methods
We recruited a consanguineous Turkish family comprising nine siblings with male triplets; two of the triplets were infertile as well as one younger infertile brother. Whole exome sequencing (WES) performed on two azoospermic brothers identified a mutation in the melanoma antigen family B4 (MAGEB4) gene which was confirmed via Sanger sequencing and then screened for on control groups and unrelated infertile subjects. The effect of the mutation on messenger RNA (mRNA) and protein levels was tested after in vitro cell transfection. Structural features of MAGEB4 were predicted throughout the conserved MAGE domain.
Results
The novel single-base substitution (c.1041A>T) in the X-linked MAGEB4 gene was identified as a no-stop mutation. The mutation is predicted to add 24 amino acids to the C-terminus of MAGEB4. Our functional studies were unable to detect any effect either on mRNA stability, intracellular localization of the protein, or the ability to homodimerize/heterodimerize with other MAGE proteins. We thus hypothesize that these additional amino acids may affect the proper protein interactions with MAGEB4 partners.
Conclusion
The whole exome analysis of a consanguineous Turkish family revealed MAGEB4 as a possible new X-linked cause of inherited male infertility. This study provides the first clue to the physiological function of a MAGE protein.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-017-0900-z) contains supplementary material, which is available to authorized users.
Keywords: Whole exome sequencing, MAGEB4, No-stop mutations, Male infertility, Azoospermia, Oligozoospermia
Introduction
Infertility affects about 7% of men worldwide; among these, a causal diagnosis is known for 60% meaning that all the other patients remain idiopathic [1]. Genetic defects could explain the cause of infertility in such men.
Among infertile males, 10 to 15% will be diagnosed with azoospermia [2] and 35% with oligoozoospermia (with variable severity) [3]. Numerical and structural chromosomal abnormalities are known genetic causes of azoospermic and oligospermic cases; approximately 14% of azoospermic men and 5% of oligozoospermic men carry chromosomal abnormalities [4]. The most frequent numerical chromosome aberration is Klinefelter’s syndrome, with an extra X chromosome (47,XXY) which is found in 14% of azoospermic men [5, 6]. Yq microdeletions are among the most frequent structural chromosomal causes of male infertility, involving the loci named azoospermia factor (AZF), and are found in about 10% of men with oligozoospermia and in up to 15% of azoospermic patients [7, 8]. Consequently, a genetic diagnosis is possible for only a few patients.
The mouse model has illustrated the complexity of spermatogenesis with concerted action of more than 2300 genes [9]. The analysis of the knockout (KO) mice for some of these genes has pinpointed their function in spermatogenesis, providing insights into some of the molecular mechanisms involved in the appearance of azoospermia and oligozoospermia [10]. Therefore, it is likely that most “idiopathic” human forms may have a genetic origin. So far, only a few human genes have been identified as responsible, when mutated, for azoospermia or oligozoospermia such as DAX1, NPAS2, NR5A1, SOHLH1, SYCE1, TAF4B, TEX11, TEX15, WT1, and ZMYND15 [11–20].
By analogy to the Y chromosome, the X chromosome is interesting in view of studying male infertility. Indeed, at the beginning of the millennium, Wang and colleagues identified 25 genes in mice that were expressed only in spermatogonia but not in somatic cells; 40% of them mapped to the X chromosome, which suggested a predominant role of these genes in mammalian spermatogenesis [21]. For some of these genes, KO mouse lines have been created and analyzed, confirming their function in spermatogenesis [22–24]. So far, few genes on the X chromosome have been studied in infertile men [25–34].
Recently, an X-linked gene, namely TEX11, was identified in which mutations are responsible for an azoospermic phenotype [17, 18]. The predominant phenotype of meiotic arrest observed in humans closely recapitulated the phenotype observed in the KO mouse model. [35]. Related mutations, ∼15% in patients with meiotic arrest and ∼1–2% among all azoospermic patients [17, 18], are the most frequently found in these conditions in addition to Yq microdeletions. Similarly, the potential role of X-linked RHOX gene cluster in human was studied. Two mutations in RHOXF1 and four in RHOXF2/2B in patients with severe oligozoospermia were revealed, which provide evidence of impaired function of RHOX genes in human male infertility [34].
Studying a consanguineous Turkish family, we identified melanoma antigen family B4 (MAGEB4, MIM 300153) as a new X-linked gene involved in an inherited male infertility. Indeed, a no-stop mutation was found to co-segregate with the non-obstructive azoospermia (NOA) and oligozoospermia phenotype in the family. This novel mutation (c.1041A>T, p.*347Cysext*24) substitutes the stop codon with a cysteine residue, potentially adding 24 amino acids to the C-terminus of MAGEB4. With the exception of a few functional studies on cancer cells, nothing is known about the physiological role of the MAGE gene family. This study provides the first clue to a physiological function of a MAGE protein.
Materials and methods
Patients and controls
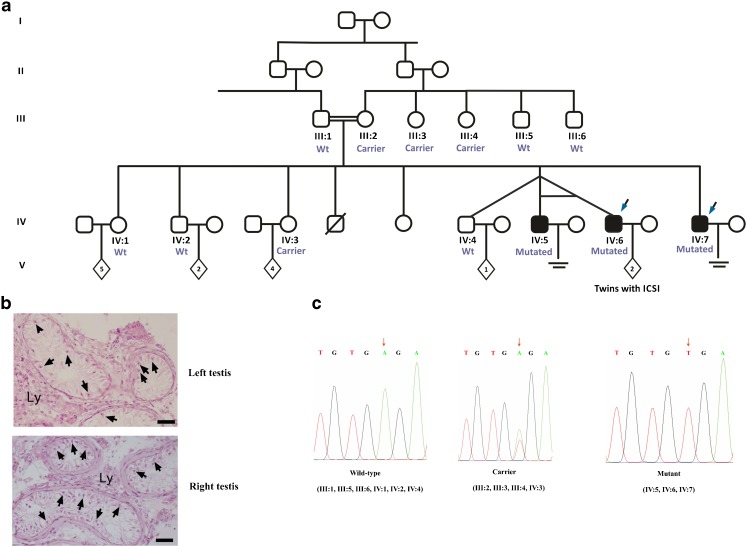
The family comprised five brothers; three infertile brothers, two of them being born of a triplet pregnancy, and two fertile brothers, including the one from the triplet pregnancy. All affected brothers have a normal karyotype, and no Yq microdeletions were identified. The two fertile brothers fathered two and one child, respectively, while two fertile sisters mothered four and five children, respectively. The mother (III:2) has four siblings; two males and two females, all have children (Fig. 1a).
Fig. 1.
Consanguineous Turkish family with nine siblings comprising male triplets. One male sibling died before the age of 2; the cause of death is unknown. IV:5 and IV:6 were monozygotic twins in the triplet. a The segregation of mutation c.1041A>T, p.*347Cysext*24 in the family. Whole exome sequencing was performed on two affected brothers indicated with blue arrows. b Testicular histology of patient IV:7. Representative sections of human left and right testis biopsies displaying absence of germinal cells without histological impairment of Sertoli and Leydig cells. Arrows indicate Sertoli cells. Ly Leydig cells. Bars represent 150 μm in length. c Plots of results from Sanger sequencing for mutant, carrier, and wild-type samples. Allele in question is indicated by orange arrow. Mutation causes stop codon (TGA) change into codon for cysteine (TGT)
A total of 108 unrelated, infertile Turkish men [88 NOA, 20 severe oligospermia (SO)] and 107 fertile Turkish males who fathered at least one child naturally were also included in the study. All semen analyses were performed according to current WHO guidelines [36].
In order to enlarge the patient cohort, we screened for the MAGEB4 mutation in 67 French patients [37 azoospermia, 30 oligozoospermia], 325 NOA patients with German origin, and 187 normozoospermic German men.
NOA cases were selected based on two spermiograms, and no spermatozoa were detected after concentration. Similarly, SO cases were selected with a threshold of <1 × 106 spermatozoa/ml after semen analysis. Exclusion criteria were patients with monolateral or bilateral cryptorchidism, varicocele, previous testis trauma, mixed azoospermia (obstructive associated with non-obstructive), recurrent infections, iatrogenic infertility, hypogonadotrophic hypogonadism, karyotype anomalies, or Yq microdeletions.
DNA preparation
Saliva samples were used as a DNA source from family members, all Turkish controls and SO cases, whereas blood samples were collected from individual Turkish NOA cases after obtaining written consent. Genomic DNA was extracted from saliva using Oragene DNA Self-Collection Kit (DNA Genotek, Ottawa, Canada) or from peripheral blood using IPrep™ Pure Link® gDNA Blood Kit (Invitrogen, CA, USA), according to the manufacturer’s instructions. Extracted DNA samples were obtained for French patients, whereas mutation screening was performed in Germany for their patients by the team of Dr. Frank Tüttelmann on DNA extracted from peripheral blood.
Whole exome sequencing and data processing and analysis
Whole exome sequencing of patients IV:6 and IV:7 (Fig. 1a) was performed as described previously [19]. Briefly, approximately 1 μg DNA was sheared to 150–200 bp with Covaris Sonolab v4.3.3 (Covaris, Woburn, MA, USA). Fragments were subjected to library preparation; for each sample to be sequenced, an individual indexed library was prepared. DNA libraries were then enriched (SureSelectXT2 Target Enrichment System, Agilent Technologies, CA, USA) and sequenced with Illumina Hiseq 2500 following Illumina’s instructions. Image analysis and base calling were performed using CASAVA v1.8.2 (Illumina), and detected variants were ranked by VaRank software [37].
Mutation screening
Candidate variants from whole exome sequencing analysis and their segregation in the family were confirmed by polymerase chain reaction (PCR) and Sanger sequencing. For the variant that showed matched segregation with the phenotype in the family, the absence of variation in healthy matched Turkish and German controls was confirmed through Sanger sequencing. All exons and exon/intron boundaries of genes of interest were sequenced for validating variants as well as looking for other mutations in a group of infertile Turkish, French, and German men with similar phenotypes. Turkish and French patients were sequenced via Sanger sequencing in our lab, while German patients were analyzed via Sanger sequencing by the team of Dr. Tüttelmann in Germany. Amplification conditions and all primers are listed in supplementary data (Tables S.1A and S.1B). DNA amplicons were purified, and double-strand sequencing of each DNA fragment were performed.
RT-PCR and immunohistology
RNA samples, RNA extraction, and complementary DNA (cDNA) synthesis were done as described previously [19]. Amplification conditions and primers for RT-PCR listed in supplementary data Table S.1C. Thirty cycles of RT-PCR were followed by gel electrophoresis and ethidium bromide staining.
For immunohistochemical evaluation, human testicular tissue samples were obtained from adult patients undergoing a vasectomy reversal in the fertility center of the UZ Brussels. During surgery, a testicular tissue sample measuring about 50 mm3 was taken to evaluate the spermatogenic status. A small piece of this testicular sample was used for research purposes after written informed consent. Prepubertal tissue was obtained from a patient who underwent testicular tissue banking as part of a fertility reservation program. Slides for immunohistological evaluation were prepared as described before [19]. MAGEB4 primary antibodies (1/50; anti-MAGEB4-antibody, ab60048 GR37013-1, Abcam, Cambridge, UK) were used for samples, and no primary antibody was applied to the negative control.
After three wash steps, sections were incubated with a peroxidase-labeled secondary antibody (Dako Real Envision Detection System; K5007; Dako, Heverlee, Belgium) for 1 h at room temperature. After washing and visualization with 3,3′-diaminobenzidine (DAB, Dako RealTM EnvisionTM Envision system), slides were counterstained with hematoxylin.
Construction of expression plasmids
DNA from III:1 and IV:7 was used as a template for wild-type and mutant constructions by PCR. Three primers (supplementary data Table S.2) were designed to generate wild-type (WT) and mutated (MT) constructs. Sequences of both constructs were confirmed via Sanger sequencing. The PCR products were first cloned into the TOPO vector, and the constructs were verified by DNA sequencing.
WT and mutant fragments were amplified through PCR by using forward and reverse primer (Supplementary data Table S.3). DsRed-Monomer N1 vector (BD Bioscience) was linearized by HindIII and AflII digestion. Amplified fragments were cloned into the linearized vector by 1 cycle of circular polymerase extension cloning (CPEC) reaction as previously described [38]. Briefly, 200 ng of the linear vector was mixed with amplified fragments at equal molar ratio in a 20 μl volume containing Phusion High Fidelity reaction mixture (ThermoScientific). A short denaturation step (98 °C, 30 s) was utilized to denature double-stranded insert and linear vector, followed by annealing step at 55 °C for 30 s, and then, polymerase extension was performed for 15 s per kilobase according to the length of the longest piece. Reaction products were digested with DpnI, and then, the mixture was transfected into XL-Blue strain for protein production.
MAGEB4 wild-type and mutant cDNAs were cloned in frame either with an HA or a Flag N-terminal tag, respectively, into pcDNA3. PCMV6 MAGEA3-Myc Flag was purchased from Origene (Rockville, USA).
Cell cultures and transfections
Messenger RNA (mRNA) stability and cellular localization of wild-type and mutant forms of MAGEB4 protein were examined in HeLa and COS7 cells, respectively. A detailed protocol is given in supplementary data method S.1.
Alignment and structure prediction
The C-terminal end of the human MAGEA3 (UniProt AC P43357) and wild-type and mutant human MAGEB4 (UniProt AC O15481) have been aligned with the sequence corresponding to the crystal structure of a MAGEA3 domain (PDB 4V0P). The Jalview software (www.jalview.org) was used for sequence alignment and analysis of the sequence conservation. Secondary structure of the C-terminal end and mutant extension was predicted according to the software package PHD (https://npsa-prabi.ibcp.fr) [39, 40]. The crystal structure representations were drawn in the Pymol software (https://www.pymol.org).
Results
Desription of patients
The family pedigree is given in Fig. 1a. The parents of the index patient are first-degree cousins. The index patient is the youngest brother (IV:7); he had a microtesticular sperm extraction (TESE) operation, and histopathology defined the first biopsy material as Sertoli cell only (Fig. 1b). In December 2014, a second biopsy resulted in a retrieval of a limited number of spermatozoa that were frozen for further intracytoplasmic sperm injection (ICSI) treatment.
Among the triplets, the two infertile brothers (IV:5, IV:6) are monozygotic twins. Patient IV:5 has severe oligozoospermia (<1 × 106 spermatozoa/ml); three ICSI attempts were performed; however, no pregnancy was obtained. Sperm morphology (1% morphologically normal spermatozoa) and embryo quality were low. Patient IV:6 was diagnosed as NOA. A TESE allowed the recovery of sperm during the first in vitro fertilization (IVF) cycle. Biopsy material was frozen because of the poor hormonal response during the ovarian stimulation of the female partner. ICSI was applied in the next cycle with thawed sperm, and pregnancy was achieved, resulting in the birth of healthy twins.
Exome sequencing data analysis
Since IV:5 and IV:6 are monozygotic twins, we decided to perform whole exome sequencing on the DNA of patient IV:6 and IV:7.
A total of 9.8 and 9.2 Gb of DNA sequence were generated for samples IV:6 and IV:7, respectively. More than 74% of the target exome in both cases were represented with greater than 40-fold coverage (Supplementary data Table S.4). Sequence data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI) and the Centre for Genomic Regulation (CRG), under accession number EGAS00001002272.
Because of the loop of consanguinity in the family, we focused our first analysis on homozygous variants shared by both infertile brothers through the whole exome by assuming a recessive mode of inheritance, but we did not exclude possible dominant or X-linked transmission. After a first round of analysis, we also checked for heterozygous and hemizygous variants. We filtered variants according to sequence quality (depth of coverage >10), minor allele frequency (MAF <1%), validation status of the reference SNP, and novelty by comparing them to our in-house exome database. Variations were scanned for their occurrence in the two largest variation databases available: Exome Aggregation Consortium (ExAC) with 60,000 samples (http://exac.broadinstitute.org/) and NHLBI Exome Variant Server with 6503 samples (http://evs.gs.washington.edu/EVS/). Finally, variations were scanned through online expression databases (Amazonia, BioGPS, EMBL-EBI expression atlas) and the literature for a suggested role in infertility; selection criteria for candidate genes were based on testis expression and possible role in spermatogenesis as well as existence of a mouse KO model with a male sterility phenotype.
At the end of the screening process, four genes remained: two showing a frameshift (CDCP2 and SON), one showing a missense (DACH2), and one with a no-stop variation (MAGEB4) (Supplementary data Table S.5).
Mutation confirmation and segregation in the family
All family members were screened through Sanger sequencing for the four selected variations. This allowed us to eliminate CDCP2, SON, and DACH2 since these variations were found in the fertile males of the family.
The substitution (c.1041A>T, p.*347Cysext*24) at the end of coding region of MAGEB4 was identified in both NOA brothers (IV:6 and IV:7) with 22 and 48 reads, respectively, and perfectly segregated with the infertility phenotype in the family. Indeed, Sanger sequencing confirmed the segregation of the variation (Fig. 1c) with the pathology. All three infertile males were hemizygous for the mutation; the mother and two fertile sisters were carriers; the father and fertile brothers were wild type (Fig. 1a). The siblings of the mother were also checked; they are all fertile; her sisters (III:3, III:4) were carrier, while fertile brothers (III:5, III:6) were wild type. Carrier sisters and female siblings of the index patient have sons. However, they were not available for this study. MAGEB4 belongs to a group of genes in the melanoma antigen family with specific expression to germ cells and a murine homolog which has been suggested to play a role in germ cell-specific mitosis [41].
The substitution leads to the stop codon being changed into a cysteine codon (p.*347Cysext*24) and causes loss of the normal translational termination. The next in frame stop codon is located 69 nucleotides downstream and according to GENSCAN (http://genes.mit.edu/GENSCAN.html) prediction 907 base pairs upstream of the polyA signal. Therefore, the mutation could give rise to a MAGEB4 protein with 24 additional amino acids. The mutation is not listed in any database collecting human sequences or repository for polymorphisms among the human population (ExAC, Exome Variant Server (EVS), ensembl, dbSNP).
In addition of scanning online databases and because there are no specific samples with Turkish origin in databases, we scanned fertile controls matching the same geographic region and/or same ethnicity of patients. Additionally, German fertile controls were checked through our collaborator.
The mutation was not detected in 107 ethnically matched healthy Turkish and 187 German fertile controls. In order to validate possible variants and to exclude other mutations, all exon and 5-UTR/3-UTR sites were sequenced in a total of 500 patients including 108 unrelated Turkish infertile cases (88 NOA, 20 SO), 67 French infertile cases (37 NOA and 30 oligozoospermia), and 325 German NOA cases. No mutation was identified.
Expression of MAGEB4
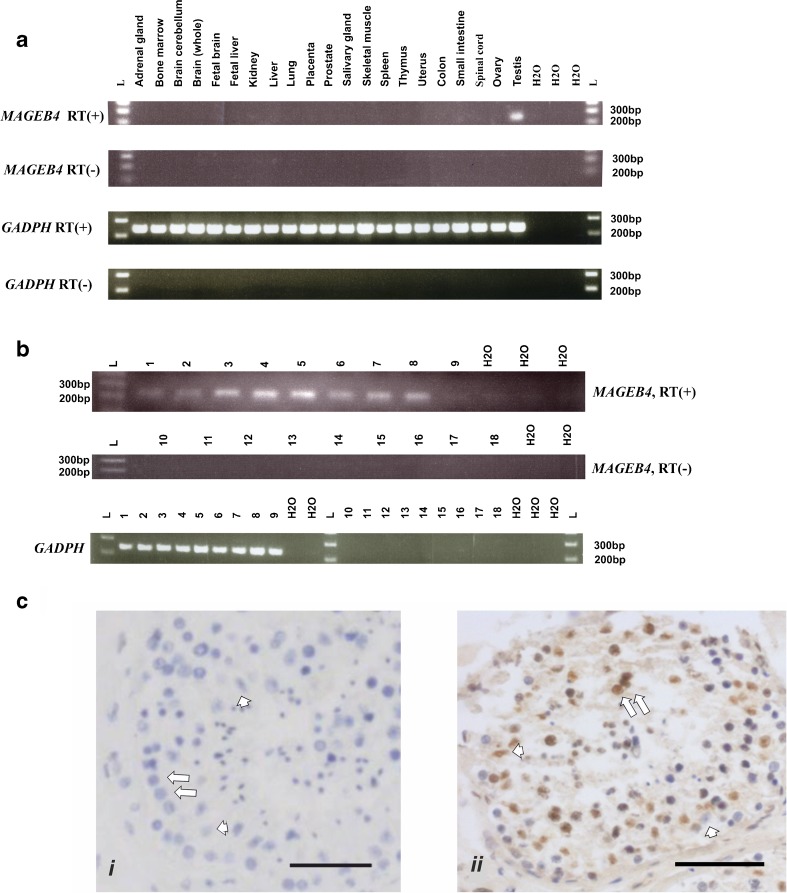
The expression profile of MAGEB4 in human tissue was studied at the mRNA and protein levels. Twenty-one total RNA samples from different organs, including testis, and nine testicular tissues derived from three Sertoli cell only, four maturation arrests at different stages, one normal, and one prepubescent patient were analyzed by RT-PCR (primers listed in supplementary data Table S1C). The results confirmed the restricted expression of MAGEB4 to the testis (Fig. 2a). With the exception of prepubertal testis, MAGEB4 expression was detectable in all human testicular tissues (Fig. 2b). This latter result was confirmed by immunohistology which showed a high level of expression in differentiating germ cells and little expression in Sertoli cells (Fig. 2c).
Fig. 2.
Expression studies for MAGEB4 at mRNA and protein levels. GADPH was used as internal control for RT-PCR; individual control without reverse transcriptase added to each sample and labeled as RT(−). a RT-PCR results of MAGEB4 and GADPH for different human tissues. b RT-PCR results of MAGEB4 and GADPH for human testicular samples with different histology. RT(+) (well nos. 1–9), RT(−) (well nos. 10–18). Sertoli cell only (samples 1–3), maturation arrest at different stages (samples 4–7), normal fertile (sample 8), prepubescent (sample 9). c Immunohistochemistry for MAGEB4. Isotype control (i). Human adult testicular tissue (ii). MAGEB4 is detected in differentiating germ cells and some Sertoli cells. Scale bars 50 μm. Germ cell (arrow). Sertoli cell (arrowhead)
Functional analysis
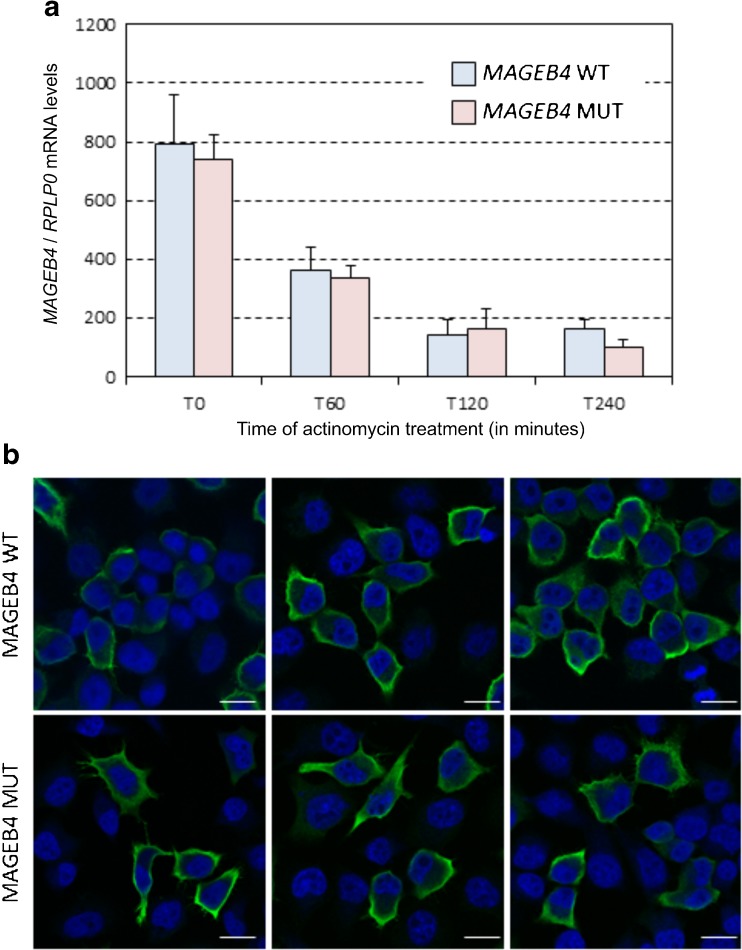
Unfortunately, we could not get access, for research purposes, to testicular biopsy samples from the affected patients, and therefore, we set up transfected cell experiments to study the effect of the variation. We first tested whether this mutation would have any effect on the stability of the messenger RNA. Quantitative RT-PCR of transfected MAGEB4 wild type and mutant indicated not only a similar level of expression of both RNA forms but also a similar rate of mRNA decay upon transcriptional inhibition by actinomycin treatment (Fig. 3a).
Fig. 3.
mRNA decay and immunofluorescence labeling for MAGEB4. a Quantitative RT-PCR of wild-type or mutant MAGEB4 mRNA expressed in HeLa cells treated with actinomycin for 1, 2, or 4 h. b Confocal images of immunofluorescence labeling of HA-tagged wild-type or mutant MAGEB4 transfected in HeLa cells. Each photo refers to a different area of the respective slides. Error bars indicate standard error of the mean (SEM). MAGEB4 is labeled in green. Nuclei are counterstained in blue with DAPI. Magnification ×400. Scale bars, 10 μm
Next, since mutation of MAGEB4 adds 24 novel amino acids to its C-terminal part, we investigated the cellular localization of a N-terminal HA-tagged wild-type and mutant MAGEB4. Both COS7 and HeLa cells were tested and gave comparable results. In transfected cells, both control and mutant proteins were expressed and localized at the cell membrane, in a pattern reminiscent of F-actin filopodia and lemellipodia structures, but with no difference between wild-type and mutant MAGEB4 (Fig. 3b).
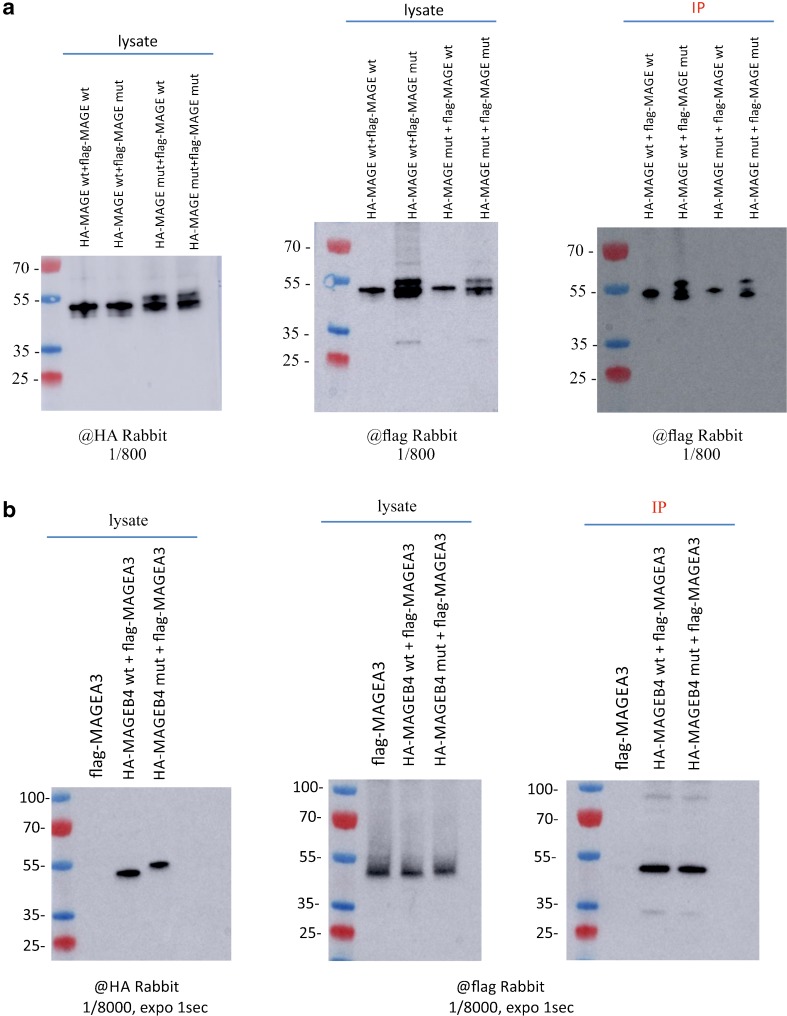
The three-dimensional X-ray structure of the MAGE domain of the human MAGEA3 protein was published recently showing the formation of a homodimer in vitro [42]. We tested the ability of wild-type and mutant MAGEB4 to form homodimers; although our immunoprecipitation studies show that the mutant MAGEB4 is a different size than the wild type, we found that MAGEB4 interacts equally well with its wild-type or mutant forms. Reciprocally, mutant MAGEB4 interacts with both wild-type and mutant MAGEB4 (Fig. 4a). We finally tested whether modifying the C-terminal part of MAGEB4 would modify its ability to interact with other proteins of the MAGE family. Co-immunoprecipitation experiments demonstrated that both wild-type and mutant MAGEB4 interact similarly with MAGEA3 (Fig. 4b).
Fig. 4.
Interaction of MAGEB4 proteins with MAGEA3. a Immunoprecipitation of Flag-tagged MAGEA3 by wild-type or mutant HA-tagged MAGEB4. b Immunoprecipitation of Flag-tagged wild-type or mutant MAGEB4 by wild-type or mutant HA-tagged MAGEB4
Structural analysis
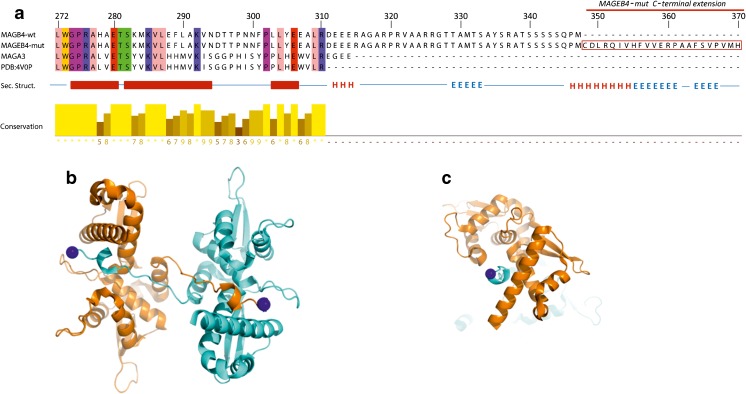
We aligned the protein sequences of MAGEB4 and MAGA3 with the mutant sequence of MAGEB4 and the sequence corresponding to the crystal structure of the MAGEA3 domain encoding residues 102–310 (PDB 4V0P) (Fig. 5a). MAGEB4 is highly homologous to MAGEA3 on the MAGE domain but has an extension of 37 residues at the C-terminal end.
Fig. 5.
Structure prediction for MAGEB4. a Sequence alignment and structural features of the C-terminal end of MAGEB4 and MAGEA3 starting at residue 272. Residues are colored according to their physicochemical properties using the Zappo coloring with 100% conservation threshold to display identities. Secondary structure elements are depicted as thick red bars for helices and red lines for loops of the crystal structure of MAGEA3. b Representation of the crystal structure of the conserved domain of MAGEA3. In the crystal, the domain of MAGEA3 (PDB 4V0P) comprising residues 102–310 (in cyan) is reaching through its C-terminal arm to a symmetry-related molecule (in orange). The last residue in the crystal structure Arg 310 is indicated by a blue sphere. c Perpendicular view of the dimer showing the C-terminal end of one monomer pointing through the other monomer
Since MAGEB4 is highly homologous to MAGEA3 throughout the conserved MAGE domain, the MAGEA3 structure spanning from residues 104–314 can be extrapolated to the MAGEB4 MAGE domain positions 272 to 347 (Fig. 5b, c). Secondary structure prediction for the C-terminal end and the additional extension in the MAGEB4 mutant indicate the presence of an α-helix preceded and followed by β-sheet regions. The C-terminal end and extension of the mutant form would extend outside the homodimeric interface shown in the crystal structure and is unlikely to disrupt this interface in agreement with our co-IP experiments.
Discussion
Despite almost four decades of IVF practice, human gametogenesis remains poorly understood. The absence of human models considerably limits knowledge of the basic mechanisms underlying human meiosis. Because of the lack of suitable in vitro models, the only effective approach to identify human genes involved in human gametogenesis is the genetic approach. Indeed, during the last decade, studying family cases of infertility or groups of infertile patients has led to a significant increase in the list of human genes involved [13–20, 34, 43–48]
We present here the analysis of a consanguineous Turkish family in which we identified, by whole exome sequencing, a single-base substitution (c.1041A>T, p.*347Cysext*24) in the X-linked MAGEB4 that segregated with the non-obstructive azoospermia and oligozoospermia phenotype.
MAGEB4 is a type I melanoma antigen family gene with specific expression to germ cells, and a murine homolog has been suggested to play a role in germ cell-specific mitosis [41]. Type I MAGE genes are encoded by three X-linked clusters including more than 45 genes [49]. Recently, it was speculated that the loss of MAGEA9, another type I MAGE gene, would affect spermatogenesis [50]. The c.1041A>T substitution changed the stop codon at position 356 to a codon for a cysteine. A new in-frame stop codon is found 69 bp downstream. So, this mutation could add 24 new amino acids to the C-terminus of MAGEB4.
No-stop mutations are very rare; there are 119 examples of mutations that occur within stop codons listed in the Human Gene Mutation Database (HGMD) 2008 update, which constitutes ∼0.2% of codon-changing mutations [51]. Because of the scarcity of this type of mutation, there is no established consensus on their consequences. In fact, “no-stop” mutations can alter protein functions in different ways. The mRNA can undergo a rapid decay, especially when no new stop codon is found before the polyA signal or over a certain distance, preventing the production of the protein [52]. The addition at the C-terminus of a stretch of amino acids can affect the stability or the folding of the protein, driving it to the proteasome for rapid degradation [53–55]. Finally, it can lead to the production of a stable protein with an altered C-terminal structure. In man, a mutation substituting the stop codon with a tyrosine residue in MSX1 has been identified, and in vitro studies showed that the mutant MSX1 could be expressed but had lost its ability to enter the nucleus [56]. There are also examples of stable proteins after no-stop mutations in other animals. In bovine protoporphyria, a no-stop mutation in the ferrochelatase gene adds 27 amino acids to the mutant protein and results in a stable, slightly larger protein in mutant animals [57]. The authors stated that this addition affects the catalytic activity of the enzyme since it occurred in the carboxyl terminus, known to play important roles in catalytic function. A no-stop mutation which resulted in the addition of 25 amino acids in the S-antigen gene was associated with late onset hereditary retinal degeneration in dogs [58]. This study also suggested that although the estimated half-life predicted localization, and instability index would not change, it is not certain that the mutant protein would fold correctly since the isoelectric point of the protein was changed. Even if the protein would fold correctly, then the mutant protein’s ability to bind appropriately to its G protein-coupled receptor, rhodopsin, would likely be impaired, and this would likely lead to the retinal degeneration phenotype [58].
Since the MAGEB4 protein is testis-restricted, for ethical reasons, we could not get access to patient’s testis samples to test these different possibilities. Therefore, we tested the effect of the mutation on MAGEB4 by in vitro cell transfection. We could show that MAGEB4 mRNA stability is not affected and that the extended protein can be produced, is stable, and is localized to the same cytoplasmic compartment as the wild-type form. This was done in a context of overexpression in cells not normally expressing MAGEB4, which may differ from the physiological level of expression and may affect the results described above. By immunoprecipitation, we also showed that mutated MAGEB4 as well as the wild-type form is able to interact with MAGEA3 and to form homodimers with itself.
Little information is available on the oligomeric state or atomic structure of MAGE proteins. The crystal structure of MAGEA3 protein indicates that the MAGE conserved core domain forms a dimer with its C-terminal end stretching on the surface of the other monomer. Our results indicate that the dimer seen in the crystal might reflect the functional dimer observed in our co-transfection experiments and that the dimer is still present despite the 37 aa-longer MAGEA3 C-terminal end. The no-stop codon mutation observed in this study adds another 24 amino acids to the C-terminus of the protein. We suggest that the MAGEB4 C-terminal tail and mutant extension would protrude from the position of Arg310 and extend outside the dimer interface (Fig. 5c). The N-terminal and C-terminal ends vary from one MAGE protein to another and might be linked to tissue and functional-specific regulations [59]. Several studies have suggested that the MAGE proteins may be involved in protein/protein interactions [49]. The mutant form, with a 24 amino acid extension of MAGEB4, does not affect the ability of the protein to homodimerize but could potentially compromise regulatory post-translational modifications and/or association with other cellular partners.
Despite the screening of 500 NOA/SO patients, we could not find any of them mutated for MAGEB4. We believe that mutation in MAGEB4 is very rare because it is part of a very large family of MAGE proteins and that there is most probable redundancy between them. Therefore, we postulate that only dominant negative mutations would give rise to a phenotype because otherwise other members of the MAGE will take over.
Although the identified mutation remains a familial one and involves only a limited number of patients, this study represents the first indication of the physiological function of the MAGEB4 protein.
Electronic supplementary material
(DOCX 32 kb).
Acknowledgements
We thank all patients for their participation and donation of samples. We would also like to thank Robert Drillien for his critical reading of the manuscript. We are grateful to the IGBMC platforms. We thank Anne-Lena Bröcher from the Institute of Human Genetics, University of Münster for her excellent technical support.
Compliance with ethical standards
Funding
The study was funded by Agence Nationale de la Recherché (ANR-11-BSV2-002 “TranspoFertil”), Fondation Maladies Rares (“High throughput sequencing and rare diseases”), and l’Agence de BioMédecine (“AMP, diagnostic prénatal et diagnostic génétique”). This work was supported by the French Centre National de la Recherche Scientifique (CNRS), Institut National de la Santé et de la Recherche Médicale (INSERM), the Ministère de l’Education Nationale et de l’Enseignement Supérieur et de la Recherche, the University of Strasbourg, and Strasbourg University Hospital. The study was also supported by the German Research Foundation (DFG) (TU298/1-2 to FT and AR).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This project has been approved by the Comité de Protection de la Personne (CPP) of Strasbourg University Hospital, France (CPP 09/40—W AC-2008-438 1W DC-2009-I 002), the “Istanbul University, Faculty of Medicine, Ethics Committee for Clinical Research Faculty of Medicine” (2012/1671-1265), and the Ethics Committee of the Medical Faculty in Münster (2010-578-f-S).
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Frank Tüttelmann and STéphane Viville are co-last authors.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-017-0900-z) contains supplementary material, which is available to authorized users.
References
- 1.Krausz C, Giachini C, Lo Giacco D, Daguin F, Chianese C, Ars E, Ruiz-Castane E, Forti G, Rossi E. High resolution X chromosome-specific array-CGH detects new CNVs in infertile males. PLoS One. 2012;7:e44887. doi: 10.1371/journal.pone.0044887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarow JP, Espeland MA, Lipshultz LI. Evaluation of the azoospermic patient. J Urol. 1989;142:62–65. doi: 10.1016/s0022-5347(17)38662-7. [DOI] [PubMed] [Google Scholar]
- 3.Baker HW. Clinical management of male infertility. In: Jameson JL, De Groot LI, editors. Endocronology. 6th. Philadelphia, PA: Elsevier Press; 2010. pp. 2556–2579. [Google Scholar]
- 4.Van Assche E, Bonduelle M, Tournaye H, Joris H, Verheyen G, Devroey P, Van Steirteghem A, Liebaers I. Cytogenetics of infertile men. Hum Reprod. 1996;11:1–24. doi: 10.1093/humrep/11.suppl_4.1. [DOI] [PubMed] [Google Scholar]
- 5.Rives N, Joly G, Machy A, Simeon N, Leclerc P, Mace B. Assessment of sex chromosome aneuploidy in sperm nuclei from 47,XXY and 46,XY/47,XXY males: comparison with fertile and infertile males with normal karyotype. Mol Hum Reprod. 2000;6:107–112. doi: 10.1093/molehr/6.2.107. [DOI] [PubMed] [Google Scholar]
- 6.Walsh TJ, Pera RR, Turek PJ. The genetics of male infertility. Semin Reprod Med. 2009;27:124–136. doi: 10.1055/s-0029-1202301. [DOI] [PubMed] [Google Scholar]
- 7.Yatsenko SA, Rajkovic A. Chromosomal causes of infertility: the story continues. In: Sermon K, Viville S, editors. Textbook of Human Reproductive Genetics. Cambridge: Cambridge University Press; 2014. pp. 98–100. [Google Scholar]
- 8.Krausz C, Hoefsloot L, Simoni M, Tüttelmann F. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions. State of the Art 2013. Andrology. 2014;2:5–19. doi: 10.1111/j.2047-2927.2013.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hotaling J, Carrell DT. Clinical genetic testing for male factor infertility: current applications and future directions. Andrology. 2012;2:339–350. doi: 10.1111/j.2047-2927.2014.00200.x. [DOI] [PubMed] [Google Scholar]
- 10.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mou L, Xie N, Yang L, Liu Y, Diao R, Cai Z, Li H, Gui Y. A novel mutation of DAX-1 associated with secretory azoospermia. PLoS One. 2015;10:e0133997. doi: 10.1371/journal.pone.0133997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramasamy R, Bakircioglu ME, Cengiz C, Karaca E, Scovell J, Jhangiani SN, Akdemir ZC, Bainbridge M, Yu Y, Huff C, Gibbs RA, Lupski JR, Lamb DJ. Whole-exome sequencing identifies novel homozygous mutation in NPAS2 in family with nonobstructive azoospermia. Fertil Steril. 2015;104:286–291. doi: 10.1016/j.fertnstert.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashamboo A, Ferraz-de-Souza B, Lourenco D, Lin L, Sebire NJ, Montjean D, Bignon-Topalovic J, Mandelbaum J, Siffroi JP, Christin-Maitre S, Radhakrishna U, Rouba H, Ravel C, Seeler J, Achermann JC, McElreavey K. Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am J Hum Genet. 2010;87:505–512. doi: 10.1016/j.ajhg.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi Y, Jeon S, Choi M, Lee MH, Park M, Lee DR, Jun KY, Kwon Y, Lee OH, Song SH, Kim JY, Lee KA, Yoon TK, Rajkovic A, Shim SH. Mutations in SOHLH1 gene associate with nonobstructive azoospermia. Hum Mutat. 2010;31:788–793. doi: 10.1002/humu.21264. [DOI] [PubMed] [Google Scholar]
- 15.Maor-Sagie E, Cinnamon Y, Yaacov B, Shaag A, Goldsmidt H, Zenvirt S, Laufer N, Richler C, Frumkin A. Deleterious mutation in SYCE1 is associated with non-obstructive azoospermia. J Assist Reprod Genet. 2015;32:887–891. doi: 10.1007/s10815-015-0445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayhan O, Balkan M, Guven A, Hazan R, Atar M, Tok A, Tolun A. Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J Med Genet. 2014;51:239–244. doi: 10.1136/jmedgenet-2013-102102. [DOI] [PubMed] [Google Scholar]
- 17.Yang F, Silber S, Leu NA, Oates RD, Marszalek JD, Skaletsky H, Brown LG, Rozen S, Page DC, Wang PJ. TEX11 is mutated in infertile men with azoospermia and regulates genome-wide recombination rates in mouse. EMBO Mol Med. 2015;7:1198–1210. doi: 10.15252/emmm.201404967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yatsenko AN, Georgiadis AP, Ropke A, Berman AJ, Jaffe T, Olszewska M, Westernstroer B, Sanfilippo J, Kurpisz M, Rajkovic A, Yatsenko SA, Kliesch S, Schlatt S, Tuttelmann F. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med. 2015;372:2097–2107. doi: 10.1056/NEJMoa1406192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okutman O, Muller J, Baert Y, Serdarogullari M, Gultomruk M, Piton A, Rombaut C, Benkhalifa M, Teletin M, Skory V, Bakircioglu E, Goossens E, Bahceci M, Viville S. Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum Mol Genet. 2015;24:5581–5588. doi: 10.1093/hmg/ddv290. [DOI] [PubMed] [Google Scholar]
- 20.Wang XN, Li ZS, Ren Y, Jiang T, Wang YQ, Chen M, Zhang J, Hao JX, Wang YB, Sha RN, Huang Y, Liu X, Hu JC, Sun GQ, Li HG, Xiong CL, Xie J, Jiang ZM, Cai ZM, Wang J, Huff V, Gui YT, Gao F. The Wilms tumor gene, Wt1, is critical for mouse spermatogenesis via regulation of sertoli cell polarity and is associated with non-obstructive azoospermia in humans. PLoS Genet. 2013;9:e1003645. doi: 10.1371/journal.pgen.1003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 22.Greenbaum MP, Yan W, Wu MH, Lin YN, Agno JE, Sharma M, Braun RE, Rajkovic A, Matzuk MM. TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci U S A. 2006;103:4982–4987. doi: 10.1073/pnas.0505123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarabay Y, Kieffer E, Teletin M, Celebi C, Van Montfoort A, Zamudio N, Achour M, El Ramy R, Gazdag E, Tropel P, Mark M, Bourc’his D, Viville S. The mammalian-specific Tex19.1 gene plays an essential role in spermatogenesis and placenta-supported development. Hum Reprod. 2013;28:2201–2214. doi: 10.1093/humrep/det129. [DOI] [PubMed] [Google Scholar]
- 24.Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol. 2008;180:673–679. doi: 10.1083/jcb.200709057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akinloye O, Gromoll J, Callies C, Nieschlag E, Simoni M. Mutation analysis of the X-chromosome linked, testis-specific TAF7L gene in spermatogenic failure. Andrologia. 2007;39:190–195. doi: 10.1111/j.1439-0272.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Park HS, Kim HH, Yun YJ, Lee DR, Lee S. Functional polymorphism in H2BFWT-5’UTR is associated with susceptibility to male infertility. J Cell Mol Med. 2009;13:1942–1951. doi: 10.1111/j.1582-4934.2009.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olesen C, Silber J, Eiberg H, Ernst E, Petersen K, Lindenberg S, Tommerup N. Mutational analysis of the human FATE gene in 144 infertile men. Hum Genet. 2003;113:195–201. doi: 10.1007/s00439-003-0974-9. [DOI] [PubMed] [Google Scholar]
- 28.Ravel C, El Houate B, Chantot S, Lourenco D, Dumaine A, Rouba H, Bandyopadahyay A, Radhakrishna U, Das B, Sengupta S, Mandelbaum J, Siffroi JP, McElreavey K. Haplotypes, mutations and male fertility: the story of the testis-specific ubiquitin protease USP26. Mol Hum Reprod. 2006;12:643–646. doi: 10.1093/molehr/gal063. [DOI] [PubMed] [Google Scholar]
- 29.Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. Possible role of USP26 in patients with severely impaired spermatogenesis. Eur J Hum Genet. 2005;13:336–340. doi: 10.1038/sj.ejhg.5201335. [DOI] [PubMed] [Google Scholar]
- 30.Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. Alterations of the USP26 gene in Caucasian men. Int J Androl. 2006;29:614–617. doi: 10.1111/j.1365-2605.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 31.Stouffs K, Tournaye H, Van der Elst J, Liebaers I, Lissens W. Is there a role for the nuclear export factor 2 gene in male infertility? Fertil Steril. 2008;90:1787–1791. doi: 10.1016/j.fertnstert.2007.08.071. [DOI] [PubMed] [Google Scholar]
- 32.Stouffs K, Tournaye H, Liebaers I, Lissens W. Male infertility and the involvement of the X chromosome. Hum Reprod Update. 2009;15:623–637. doi: 10.1093/humupd/dmp023. [DOI] [PubMed] [Google Scholar]
- 33.Visser L, Westerveld GH, Xie F, van Daalen SK, van der Veen F, Lombardi MP, Repping SA. Comprehensive gene mutation screen in men with asthenozoospermia. Fertil Steril. 2011;95:1020–1024. doi: 10.1016/j.fertnstert.2010.11.067. [DOI] [PubMed] [Google Scholar]
- 34.Borgmann J, Tüttelmann F, Dworniczak B, Röpke A, Song HW, Kliesch S, Wilkinson MF, Laurentino S, Gromoll J. The human RHOX gene cluster: target genes and functional analysis of gene variants in infertile men. Hum Mol Genet 2016; Sep 15. pii: ddw313. [DOI] [PMC free article] [PubMed]
- 35.Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Höög C, McLaughlin KJ, Wang PJ. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 2008;22(5):682–691. doi: 10.1101/gad.1613608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. WHO laboratory manual for the examination and processing of human semen. 5th ed. WHO Press, 2010.
- 37.Geoffroy V, Pizot C, Redin C, Piton A, Vasli N, Stoetzel C, Blavier A, Laporte J, Muller J. VaRank: a simple and powerful tool for ranking genetic variants. Peer J. 2015;3:e796. doi: 10.7717/peerj.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 40.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 41.Osterlund C, Tohonen V, Forslund KO, Nordqvist K. Mage-b4, a novel melanoma antigen (MAGE) gene specifically expressed during germ cell differentiation. Cancer Res. 2000;60:1054–1061. [PubMed] [Google Scholar]
- 42.Newman JA, Cooper CDO, Roos AK, Aitkenhead H, Oppermann UCT, Cho HJ, Osman R, Gileadi O. Structures of two melanoma-associated antigens suggest allosteric regulation of effector binding. PLoS One. 2016;11:e0148762. doi: 10.1371/journal.pone.0148762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avenarius MR, Hildebrand MS, Zhang Y, Meyer NC, Smith LL, Kahrizi K, Najmabadi H, Smith RJ. Human male infertility caused by mutations in the CATSPER1 channel protein. Am J Hum Genet. 2009;84:505–510. doi: 10.1016/j.ajhg.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben Khelifa M, Coutton C, Zouari R, Karaouzene T, Rendu J, Bidart M, Yassine S, Pierre V, Delaroche J, Hennebicq S, Grunwald D, Escalier D, Pernet-Gallay K, Jouk PS, Thierry-Mieg N, Toure A, Arnoult C, Ray PF. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caburet S, Arboleda VA, Llano E, Overbeek PA, Barbero JL, Oka K, Harrison W, Vaiman D, Ben-Neriah Z, Garcia-Tunon I, Fellous M, Pendas AM, Veitia RA, Vilain E. Mutant cohesin in premature ovarian failure. N Engl J Med. 2014;370:943–949. doi: 10.1056/NEJMoa1309635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dam AH, Koscinski I, Kremer JA, Moutou C, Jaeger AS, Oudakker AR, Tournaye H, Charlet N, Lagier-Tourenne C, van Bokhoven H, Viville S. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet. 2007;81:813–820. doi: 10.1086/521314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dieterich K, Soto Rifo R, Faure AK, Hennebicq S, Ben Amar B, Zahi M, Perrin J, Martinez D, Sele B, Jouk PS, Ohlmann T, Rousseaux S, Lunardi J, Ray PF. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet. 2007;39:661–665. doi: 10.1038/ng2027. [DOI] [PubMed] [Google Scholar]
- 48.Koscinski I, Elinati E, Fossard C, Redin C, Muller J, Velez de la Calle J, Schmitt F, Ben Khelifa M, Ray PF, Kilani Z, Barratt CL, Viville S. DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet. 2011;88:344–350. doi: 10.1016/j.ajhg.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo Giacco D, Chianese C, Ars E, Ruiz-Castané E, Forti G, Krausz C. Recurrent X chromosome-linked deletions: discovery of new genetic factors in male infertility. J Med Genet. 2014;51:340–344. doi: 10.1136/jmedgenet-2013-101988. [DOI] [PubMed] [Google Scholar]
- 51.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamby SE, Thomas NS, Cooper DN, Chuzhanova N. A meta-analysis of single base-pair substitutions in translational termination codons (‘nonstop’ mutations) that cause human inherited disease. Hum Genomics. 2011;5:241–264. doi: 10.1186/1479-7364-5-4-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cacciottolo M, Numitone G, Aurino S, Caserta IR, Fanin M, Politano L, Minetti C, Ricci E, Piluso G, Angelini C, Nigro V. Muscular dystrophy with marked dysferlin deficiency is consistently caused by primary dysferlin gene mutations. Eur J Hum Genet. 2011;19:974–980. doi: 10.1038/ejhg.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu LL, Li XH, Han Y, Zhang DH, Gong QM, Zhang XX. A novel homozygous no-stop mutation in G6PC gene from a Chinese patient with glycogen storage disease type Ia. Gene. 2014;536:362–365. doi: 10.1016/j.gene.2013.11.059. [DOI] [PubMed] [Google Scholar]
- 55.Oegema R, Hulst JM, Theuns-Valks SD, van Unen LM, Schot R, Mancini GM, Schipper ME, de Wit MC, Sibbles BJ, de Coo IF, Nanninga V, Hofstra RM, Halley DJ, Brooks AS. Novel no-stop FLNA mutation causes multi-organ involvement in males. Am J Med Genet A. 2013;161A:2376–2384. doi: 10.1002/ajmg.a.36109. [DOI] [PubMed] [Google Scholar]
- 56.Wong SW, Liu HC, Han D, Chang HG, Zhao HS, Wang YX, Feng HL. A novel non-stop mutation in MSX1 causing autosomal dominant non-syndromic oligodontia. Mutagenesis. 2014;29:319–323. doi: 10.1093/mutage/geu019. [DOI] [PubMed] [Google Scholar]
- 57.Jenkins MM, LeBoeuf RD, Ruth GR, Bloomer JR. A novel stop codon mutation (X417L) of the ferrochelatase gene in bovine protoporphyria, a natural animal model of the human disease. Biochim Biophys Acta. 1998;1408:18–24. doi: 10.1016/S0925-4439(98)00052-0. [DOI] [PubMed] [Google Scholar]
- 58.Goldstein O, Jordan JA, Aguirre GD, Acland GM. A non-stop S-antigen gene mutation is associated with late onset hereditary retinal degeneration in dogs. Mol Vis. 2013;19:1871–1884. [PMC free article] [PubMed] [Google Scholar]
- 59.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 32 kb).