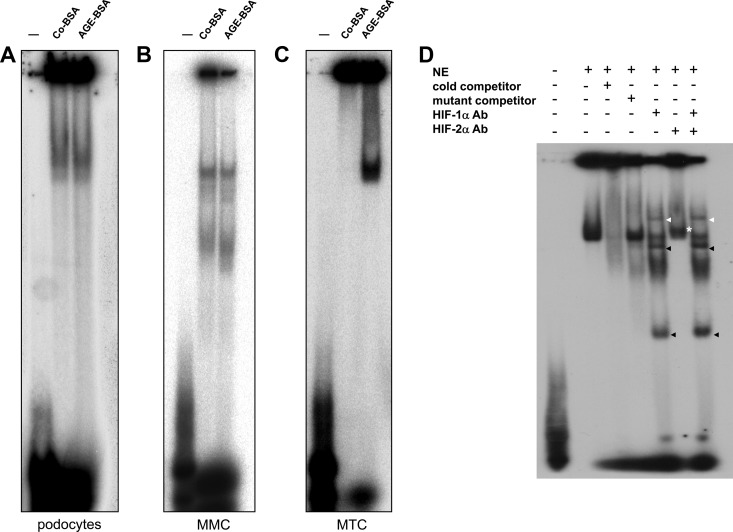
Figure 7.
A–D, Influence of AGE-BSA on HIF-1/2α transcriptional activity analyzed by EMSA. Nuclear proteins were extracted from podocytes, MMCs, and MTCs treated with Co-BSA or AGE-BSA for 24 hours and were subjected to EMSA using radioactively labeled HIF-1α double-stranded consensus oligonucleotides. Representative images of the EMSAs are shown (n = 4 independent experiments with qualitatively similar results). A, EMSA analysis of podocyte nuclear extracts (NE) from Co-BSA- or AGE-BSA-treated cells. B, EMSA analysis of MMC nuclear extracts (NE) from Co-BSA- or AGE-BSA-treated cells. C, EMSA analysis of MTC nuclear extracts (NE) from Co-BSA- or AGE-BSA-treated cells. The lane labeled with − was only loaded with the oligonucleotides without protein extracts as a negative control. A specific band is already found in podocytes and MMCs under a basal (Co-BSA) condition. In these two cell lines, AGE-BSA failed to further induce binding of transcription factors in vitro. In contrast, in MTCs (C), AGE-BSA induced a strong binding of nuclear proteins to HIF-1α double-stranded consensus oligonucleotides. D, EMSA analysis of MTC nuclear protein extracts (NE) from AGE-BSA-treated cells. The specificity of the reaction was tested by incubation of the NEs with cold competitor oligonucleotides prior to the assay or by mutant (−) oligonucleotides. In addition, the NEs were subjected to a supershift assay in which the HIF-1α and HIF-2α antibodies were added alone or together to the corresponding reaction. The HIF-1α supershift is marked with a white arrowhead, and the additional bands detected in the EMSA when the HIF-1α antibody was added to the reactions are shown with black arrowheads. The same bands occurred as well when both antibodies were added together.