Figure 2.
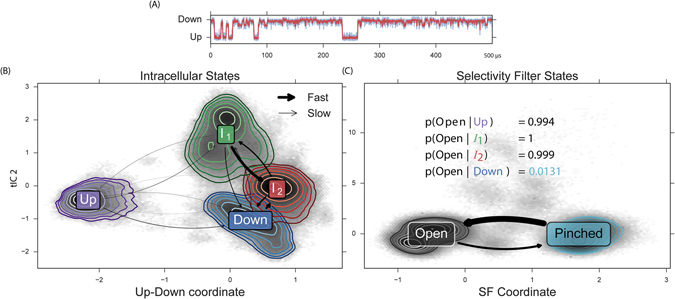
Automatic coordinate discovery using molecular dynamics data identifies an up-down coordinate from a set of high-dimensional input data. Modeling the observed dynamics by construction of an MSM permits rapid analysis of conformational dynamics. From our analysis, we discover a novel selectivity filter (SF) conformation. (A) MSM analysis permits generation of a 500 μs representative trajectory which contains transitions between up, down, and intermediate conformations, see movie in SI. (B) We find four metastable conformational macrostates, shown as contours in tIC space. Transition rates show a rapid relaxation from I 1 to I 2, and a fast exchange between I 2 and Down. (C) Automatic coordinate discovery on the selectivity filter finds a structurally distinct pinched state. Inset enumerates the SF open probability as a function of intracellular macrostate. The Up and intermediate intracellular states favor an open SF, while the Down intracellular state strongly favors a pinched SF.
