Figure 1.
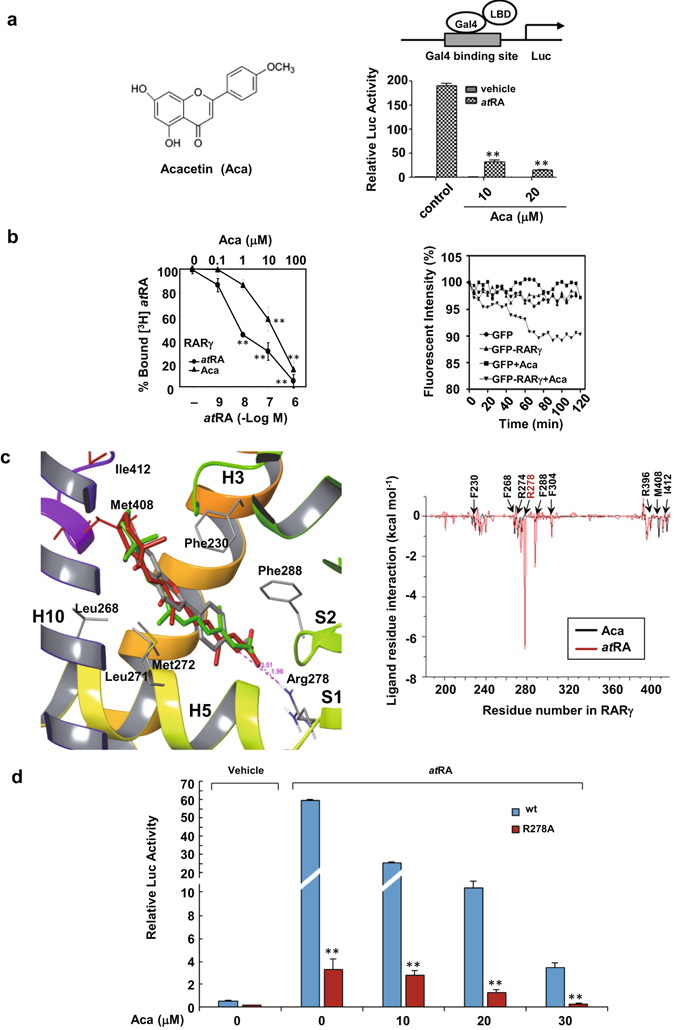
Acacetin antagonizes atRA-induced RARγ transcription and binds to RARγ. (a) The chemical structure of acacetin is indicated (left). HEK293T cells were co-transfected with pG5-luciferase reporter and pBind-RARγ LBD constructs. The cells were treated with vehicle or acacetin for 12 h in the presence or absence of 10−7 M atRA and then assayed for luciferase and β-galactosidase activities (right). (b) The purified RARγ LBD was incubated with [3H]atRA in the presence of either atRA or acacetin at an increasing concentrations. The capabilities of unlabeled atRA and acacetin to displace the radio-labeled [3H]atRA were evaluated by liquid scintillation counting after 12 h treatment (left). For cell-based assays, HepG2 cells transfected with GFP or GFP-RARγ were exposed to 10 μM acacetin or vehicle for 2 h. The images of live cells incubated at 37 °C were taken every 5 min for assaying the fluorescence intensity (right). (c) Structural comparison of acacetin (gray), 9-cis-RA (green), and atRA (red) on their interactions with the RARγLBD (left). MM-PB/SA was used to calculate the binding free energies in each residue of amino acids 180–450 in the RARγLBD that was docked with atRA and acacetin (right). (d) HEK293T cells were transfected with Gal4-RARγ/wt or Gal4-RARγ/R278A plasmids and treated with atRA (10−7 M) in the presence or absence of acacetin at different concentrations. For (a), (b) and (d), the data represent mean ± S.D. of relative luciferase activity generated in triplicate transfections in at least three independent experiments. *p < 0.05 and **p < 0.01 in (a) (acacetin + atRA vs atRA), in (b) (vs vehicle control) and in (d) (R278 vs wt).
