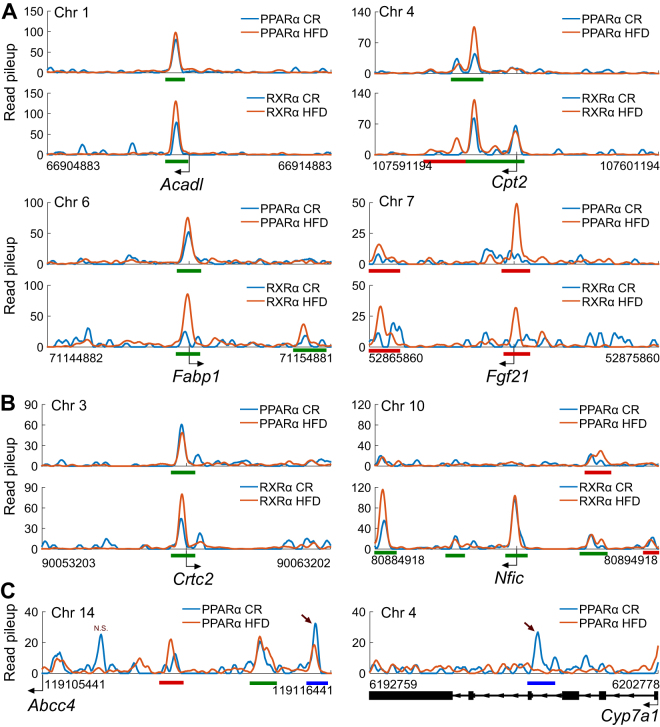
Figure 3.
ChIP-Seq of PPARα and RXRα transcription factors in CR and HFD livers reveals extensive binding near known and novel regulated genes. (A) The binding profiles (+/−5 kb gene TSS) for known PPARα and RXRα targets Acadl, Cpt2, Fabp1, and Fgf21 in CR and HFD livers are shown. (B) The binding profiles (+/−5 kb gene TSS) for novel PPARα and RXRα targets Crtc2 and Nfic in CR and HFD livers are shown. (C) The binding profiles for PPARα near the differentially expressed genes Abcc4 and Cyp7a1 (CR vs. HFD) that contain differential binding events between the same two diets in our ChIP-Seq data. Arrows indicate differential binding regions; N.S. stands for not significant. Read pileup refers to extended, normalized, and smoothed read pileup counts extracted from concatenated pools of aligned reads for the biological replicates for each factor (see Methods). Green lines indicate significantly called peaks in both CR and HFD. Red and blue lines indicate significantly called peaks in HFD and CR, respectively.