Fig. 2.
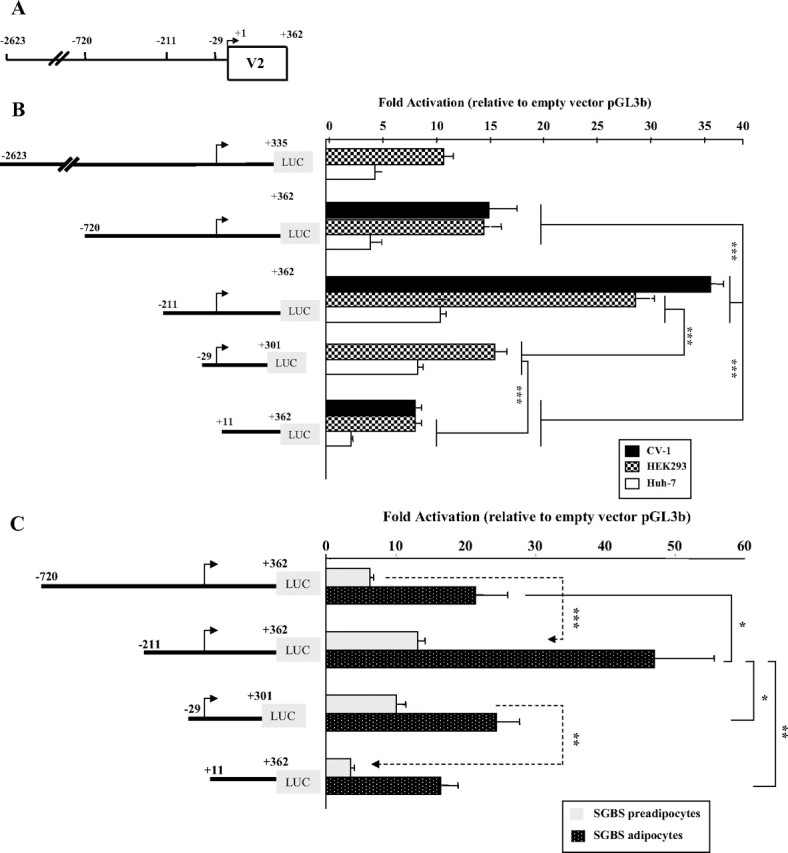
5′-Deletion analysis of the promoter activity of hGHR V2. We chose to designate the ovine 1B major TSS (T) as position +1 for our human V2 promoter construct numeration, because it represents the most 5′-cDNA end identified to date among V2 homologs (ovine 1B, bovine 1B, and mouse L2). A, A schematic diagram representing the first 362 nucleotides of the hGHR V2 exon and 2.623 kb of its 5′-flanking region. The numbering is relative to the designated major TSS (+1, indicated by arrow). 5′-Deletion promoter reporter constructs were prepared by inserting different portions of the 5′-flanking sequence of hGHR V2 into the promoterless luciferase plasmid pGL3-basic (pGL3b). B and C, These expression plasmids were transiently transfected into HEK293, CV-1 and Huh7 cells (B) and SGBS preadipocytes and mature adipocytes (C). Luciferase activity of the transfectants was normalized to β-galactosidase activity and then expressed as relative fold activation compared with the empty pGL3-b vector. The data are expressed as mean ± se; n = 3–9 experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
