Figure 2.
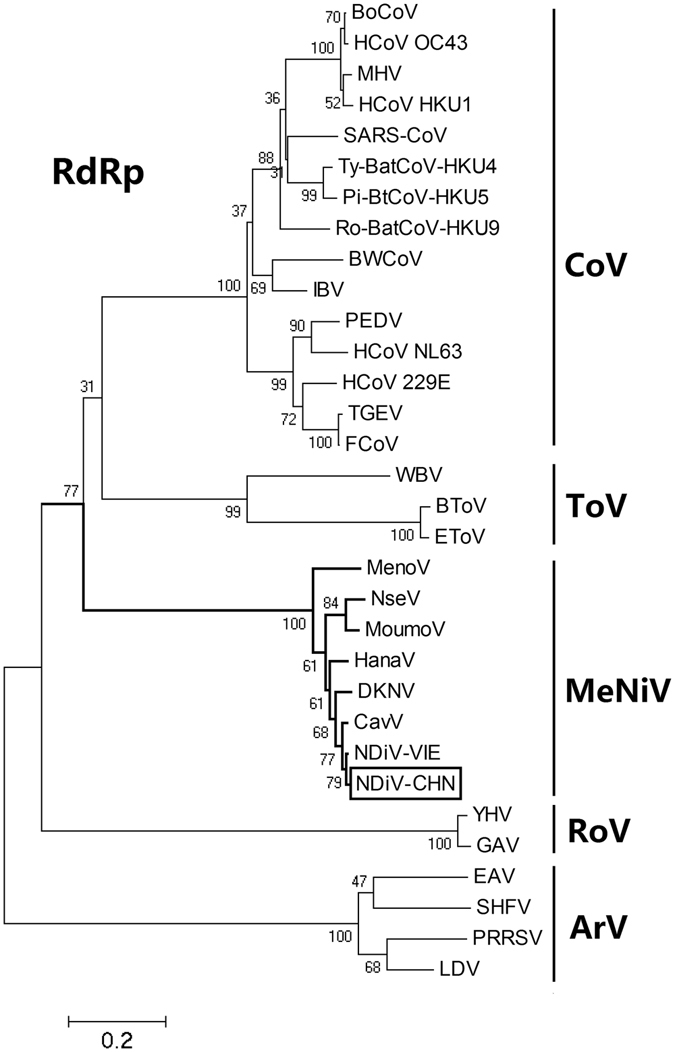
Phylogenetic analysis of nidoviruses based on the deduced amino acid sequences of RdRp domains. The phylogenetic tree was produced by the neighbor-joining (NJ) method and constructed with 1,000 bootstrap replications. The scale indicates the number of amino acid substitutions. The tree was rooted using the arterivirus branch. The GenBank/Refseq accession numbers of the nidovirus species are as follows: BoCoV, bovine coronavirus (NC_003045); HCoV OC43, human coronavirus OC43 strain ATCC VR-759 (AY585228); MHV, murine hepatitis virus strain A59 (AY700211); HCoV HKU1, human coronavirus HKU1 genotype B (AY884001); SARS-CoV, SARS coronavirus CUHK-AG03 (AY345988); Ty-BatCoV-HKU4, bat coronavirus HKU4-1 (EF065505); Pi-BtCoV-HKU5, bat coronavirus HKU5-1 (EF065509); Ro-BatCoV-HKU9, bat coronavirus HKU9-1 (EF065513); BWCoV, Beluga whale coronavirus SW1 (NC_010646); IBV, avian infectious bronchitis virus (NC_001451); PEDV, porcine epidemic diarrhea virus (NC_003436); HCoV NL63, human coronavirus NL63 isolate Amsterdam 057 (DQ445911); HCoV 229E, human coronavirus 229E (NC_002645); TGEV, TGEV Purdue P115 (DQ811788); FCoV, feline coronavirus (NC_007025); WBV, white bream virus (NC_008516); BToV, Breda virus (NC_007447); EToV, Berne virus (X52374); MenoV, Meno virus strain E9/CI/2004 (JQ957873); NseV, Nse virus strain F24/CI/2004 (JQ957874); MoumouV, Moumou virus strain C88/CI/2004 (KC768950); HanaV, Hana virus strain A4/CI/2004 (JQ957872); DKNV, Dak Nong virus (AB753015); CavV, Cavally virus isolate C79 (HM746600); NDiV-VIE, Nam Dinh virus isolate 02VN178 (DQ458789); NDiV-CHN, Nam Dinh virus isolate SZ11706Z (KF522691); YHV, yellow head virus (EU487200); GAV, gill-associated virus (NC_010306); EAV, equine arteritis virus (NC_002532); SHFV, simian hemorrhagic fever virus (NC_003092); PRRSV, porcine reproductive and respiratory syndrome virus isolate PA8 (AF176348); LDV, lactate dehydrogenase-elevating virus Plagemann strain (U15146).
