Figure 1.
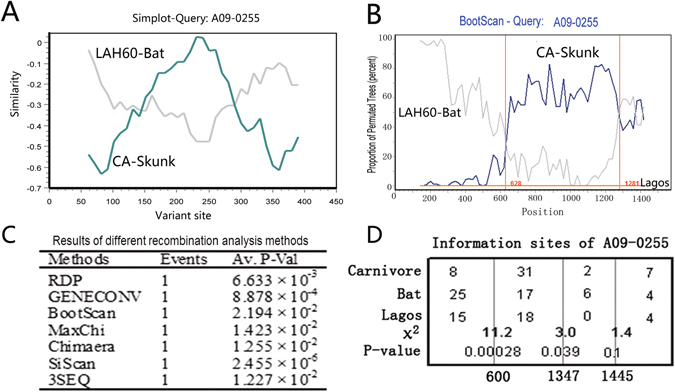
Recombination analysis of SCSKV and bat and skunk RABVs. (A) Variable site comparison of the G genes of SCSKV and carnivore and bat RABV representatives, A09-0255, CA, and LAH60. SCSKV A09-0255 was used as a query. The y-axis gives the percent identity between A09-0255 and two representatives of the parental lineages. (B) A bootscan plot of A09-0255 with a sliding window of 350 bp and a width of 20 bp using the complete G gene. The y-axis represents the percentage of permuted trees. Two Lagos bat viruses, 8619NGA and 0406SEN, were used as an outgroup. Analyses were performed using Simplot software. Vertical lines represent crossover sites. (C) Statistical analysis of the recombination event in the A09-0255 G gene using RDP 3.0. P-values of seven statistical methods implemented in RDP 3.0 to identify recombination are listed. (D) Analysis of informative sites for identification of potential recombination breakpoints. Three sites with maximum χ2 values are indicated by three vertical lines. The χ2 value and P-value of Fisher’s exact test are shown near the vertical lines. The Lagos virus isolate 8619NGA is used as the outgroup.
