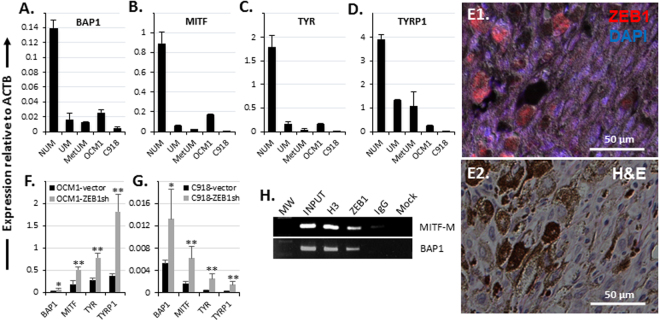
Figure 3.
ZEB1 coordinates other EMT-TFs in transcriptional regulation of EMT molecules. Transcriptional expression levels of (A) the epithelial marker E-cadherin (CDH1), (B) the mesenchymal marker N-cadherin (CDH2) detected by qPCR in NUM, UM, MetUM, OCM1 and C918 were compared. The presence of (C) CDH1 in the choroidal tissue but not in the tumor, and (D) more CDH2 in the tumor than in the choroid was detected by immunohistochemistry. (E) ZEB1 knockdown in both C918 and OCM1 and ZEB1 overexpression in OCM1 were validated by Western blots (WB). Knockdown of ZEB1 resulted in CDH1 accumulation in C918. Real-time qPCR analyses of the effects of (F) ZEB1 knockdown on the expression of the other EMT-TF genes including (G) ZEB2, (H) ID2, (I) SNAI1, (J) SNAI2, (K) TWIST1, and EMT marker genes including (L) CDH1, and (M) CDH2. (N) Chromatin immune-precipitated (ChIP) by ZEB1 antiserum (ZEB1) was utilized to detect the binding of ZEB1 to the EMT-TF genes SNAI1, TWIST1 and ID2, and to the EMT marker genes CDH1 and CDH2. Molecular weight (MW) marks were loaded on the far left lane for validation of ChIP-PCR amplicon size, followed by 10x diluted chromatin sample before immuneprecipitation (INPUT). Pan histone 3 (H3) and pre-immune serum (IgG) were served as a positive and a negative control, respectively, whereas any remaining chromatin after passing through the binding beads without any serum was served as a background control (Mock). Mean (M) ± standard deviation (SD). ‘*’ Indicates Student’s t-test p ≤ 0.05, ‘**’ Indicates Student’s t-test p ≤ 0.01.