Abstract
Background:
Neoadjuvant chemotherapy for patients with high-grade osteosarcoma has highly improved the clinical survival. However, the prognostic and predictive role of P16 expression after neoadjuvant chemotherapy remains unclear. We first determined whether P16 expression can become a potential prognostic and predictive biomarker in high-grade osteosarcoma.
Methods:
This meta-analysis was conducted based on the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guideline. Eligible studies were pooled and the overall odds ratios (ORs) and hazard ratios (HRs) with the corresponding 95% confidence intervals (95% CIs) were calculated in this analysis.
Results:
Four studies involving a total of 527 patients with high-grade osteosarcoma receiving neoadjuvant chemotherapy were identified. We did not find that P16 expression was correlated with sex status, histologic subtype, and tumor site (P > .1). P16 expression was found to be significantly associated with a “good” response to neoadjuvant chemotherapy (OR = 4.69, P < .001). A significant relationship was observed between p16 expression and pathologic complete response after neoadjuvant chemotherapy using multivariate analysis (OR = 9.63, P = .001). The expression of the P16 was not associated with clinical outcomes in overall survival (OS) and disease-free survival (DFS) by multivariate analysis (OS: P = .448; DFS: P = .263).
Conclusions:
The use of P16 expression could become a promising predictive biomarker of the response to neoadjuvant chemotherapy in the white population with high-grade osteosarcoma. However, it was not correlated with the prognosis of patients in OS and DFS. More clinical researches are very essential in Asians in the future.
Keywords: expression, high-grade osteosarcoma, neoadjuvant chemotherapy, P16, prognosis
1. Introduction
Osteosarcoma is the most common malignant neoplasm that occurred in primary bone in children and adolescents, arising from the mesenchymal tissues by tumor cells.[1,2] Its incidence is about 2 to 3 per million.[3] In the 1950s, chemotherapy was not introduced as part of optional therapies, and a 5-year survival rate is only 10% to 20%.[4,5] However, with the aid of adjuvant/neoadjuvant chemotherapy as well as advanced surgery, the clinical outcome of osteosarcoma has been significantly improved, with the long-term survival rates from 60% to 80%.[6–9] The treatment of patients with osteosarcoma with local recurrence and clinically evident metastasis still has an unsatisfactory prognosis (a 5-year survival rate of <20%).[6,10,11]
A large number of studies have suggested that the genetic and molecular mechanisms are involved in the pathogenesis, progression, and prognosis of osteosarcoma.[12–15] Localized on chromosome 9p21, the human P16 protein, a cyclin-dependent kinase inhibitor 2A (CDKN2A), encompassing 3 exons and 2 introns, participates in the regulation of cell cycle by preventing cell progression in the G1-to-S phase.[16,17] P16 expression is shown to play an impartment in cancer occurrence, progression, and prognosis.[18–20] Some studies have indicated that the altered expression of P16 was correlated with the pathogenesis and development of human osteosarcoma.[21,22] Some reports demonstrated that the association was found between the expression of P16 and the survival of osteosarcoma patients, but others found no association.[23,24]
However, the significance of P16 protein expression has not been evaluated in relation to the clinical outcome and chemotherapy response using multivariate analysis in osteosarcoma. Based on an individual study with sample sizes, we performed this meta-analysis in a large population with high-grade osteosarcoma after neoadjuvant chemotherapy to determine whether the immunohistochemical expression of p16 protein had a promising prognostic and predictive role.
2. Materials and methods
2.1. Literature search strategy
We searched a range of online digital databases to identify relevant articles published before October 19th, 2016, including the PubMed, EMBASE, EBSCO, and Cochrane Library. The following keywords and search terms used included: (osteosarcoma or osteogenic sarcoma) AND (P16 OR cyclin-dependent kinase inhibitor 2A OR CDKN2A OR INK4A) AND expression. We manually scanned the citations of the included studies to achieve other potential publications.
2.2. Selection of studies
Articles of the eligibility had to satisfy the following inclusion criteria: the patients were clinically diagnosed with osteosarcoma by histopathological examination; study of tissue specimens available receiving neoadjuvant chemotherapy was included in the current analysis; the expression of P16 protein were performed by immunohistochemistry (IHC); the studies had sufficient information to determine the association of P16 expression with the clinicopathological characteristics of patients with osteosarcoma; studies provide the hazard ratio (HR) value of P16 expression for overall survival (OS) or disease-free survival (DFS). To avoid duplicated publications from overlapping samples, only most recent article or the most complete article with more information was selected in our study.
2.3. Ethical review
This meta-analysis was not primary research involving human samples, but it was a secondary analysis regarding human subject data published in the public domain.
2.4. Data extraction
Data were extracted from eligible studies based on the selection criteria, including first author's last name, year of publication, ethnicity, number of study subjects, age, detection method (IHC), cutoff value (positivity), frequency of the P16 expression, sex status, tumor histology, tumor location, pathologic response to neoadjuvant chemotherapy, and effects on clinical outcomes (OS or DFS).
2.5. Statistical analysis
This meta-analysis was carried out using Stata software (version 12.0, Stata Corporation, College Station, TX). The pooled odds ratios (ORs) and the corresponding 95% confidence intervals (95% CIs) were calculated to estimate the relationship of P16 expression with the clinicopathological features of cancer, such as sex status, tumor histotypes, tumor location, and pathologic complete response. The pooled HRs with their 95% CIs were also analyzed to determine whether the expression of P16 protein was correlated with OS and DFS in patients with high-grade osteosarcoma. Heterogeneity among the studies was measured using the χ2 test and Q statistic.[25] A P value of <.1 for the Cochrane Q test was considered to be statistically significant heterogeneity. For the results with >2 studies, we performed a sensitivity analysis to estimate the influence of an individual study on the recalculated OR and heterogeneity by omitting one study.[26] The random-effects model was applied in the current analysis.[27,28]
3. Results
3.1. Characteristics of included studies
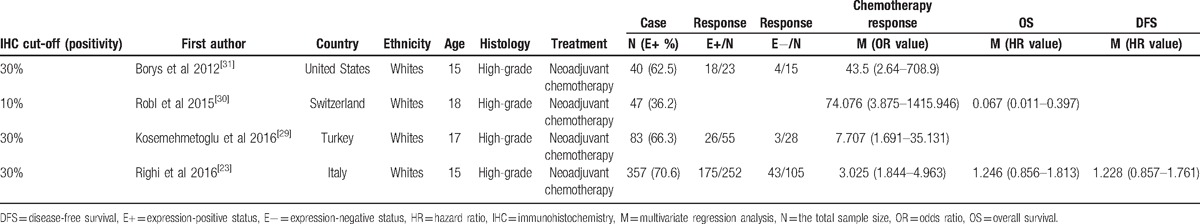
As shown in Figure 1, a total of 261 potentially relevant publications were initially retrieved through the above-mentioned databases (PubMed, EMBASE, EBSCO, and Cochrane Library) and a manual search. According to the inclusion section, after carefully reading the texts, the current meta-analysis was performed on the final 4 retrospective studies by IHC method, including 527 patients with high-grade osteosarcoma.[23,29–31] All patients belonged to the white population. The basic characteristics of 4 available studies were listed in Table 1.
Figure 1.
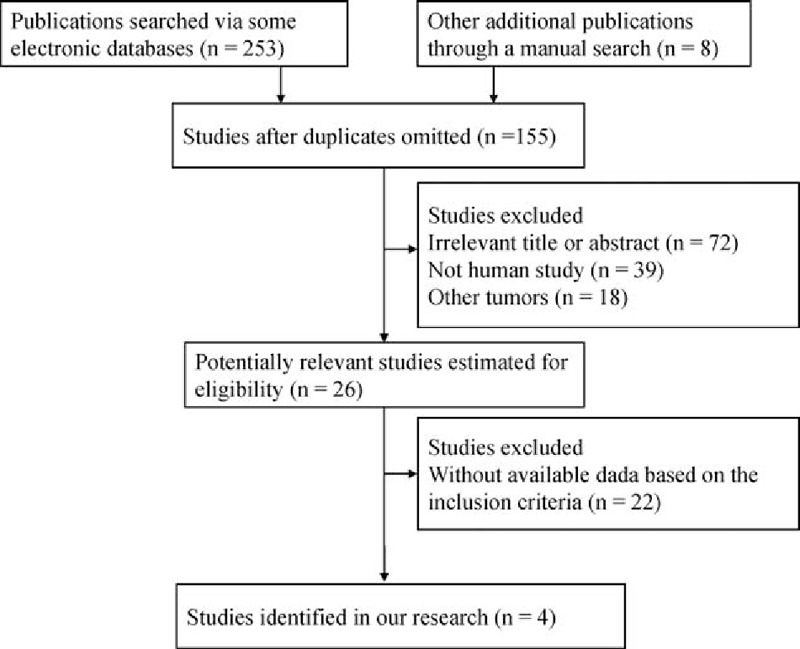
Flow diagram of study selection procedure.
Table 1.
General characteristics of the eligible studies.
3.2. Correlation between the expression of P16 protein and sex status, tumor histology, and location
We determined whether the expression of P16 protein was associated with the clinicopathological characteristics, such as sex status, tumor histopypes, and tumor site (Fig. 2).
Figure 2.
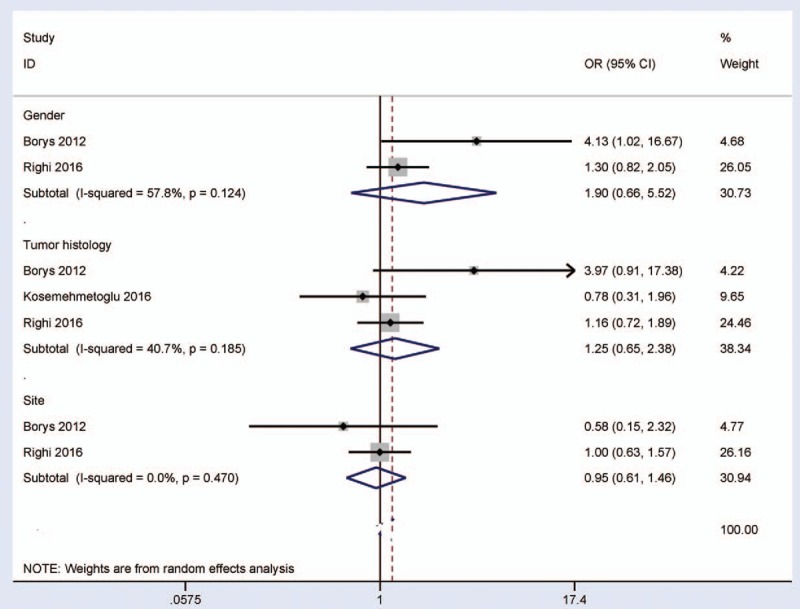
Forest plot for the associations of P16 protein expression with clinicopathological features in high-grade osteosarcoma, including sex status, tumor histology, and tumor site.
The pooled OR from 2 studies involving 219 males and females’ high-grade osteosarcoma, indicating that P16 protein expression was not correlated with sex status (OR = 1.90, 95% CI = 0.66–5.52, P = .236).
The overall OR showed that no significant correlation was found between P16 protein expression and histological subtype (OR = 1.25, 95% CI = 0.65–2.38, P = .508), including 3 studies with 313 patients with the osteoblastic type and 162 patients with nonosteoblastic type.
The pooled OR from 2 studies demonstrated that P16 protein expression was not correlated with tumor location (OR = 0.95, 95% CI = 0.61–1.46, P = .801), including the femur in 194 cases and the non-femur in 200 cases.
3.3. Correlation between the expression of P16 protein and pathologic complete response
When P16-expressing tumors were compared to P16-nonexpressing tumors, a significant correlation was observed between p16 expression and pathologic complete response to neoadjuvant chemotherapy (OR = 4.69, 95% CI = 2.36–9.31, P < .001) (Fig. 3), including 330 P16-expressing tumors and 148 P16-nonexpressing tumors. We further extracted the original results of multivariate logistic regression analysis to examine whether the expression of P16 protein can become a potential predictor of “good” neoadjuvant chemotherapy response. The result showed that P16 expression was significantly correlated with tumor response after neoadjuvant chemotherapy (OR = 9.63, 95% CI = 2.39–38.85, P = .001), including 4 studies with 527 high-grade osteosarcoma patients (Fig. 4).
Figure 3.
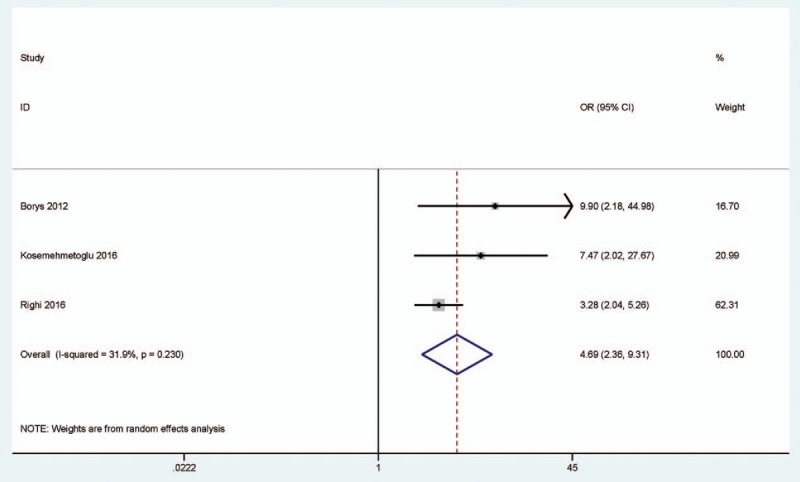
Forest plot for the association between P16 protein expression and pathologic complete response to neoadjuvant chemotherapy in P16-expressing tumors vs. P16-nonexpressing tumors.
Figure 4.
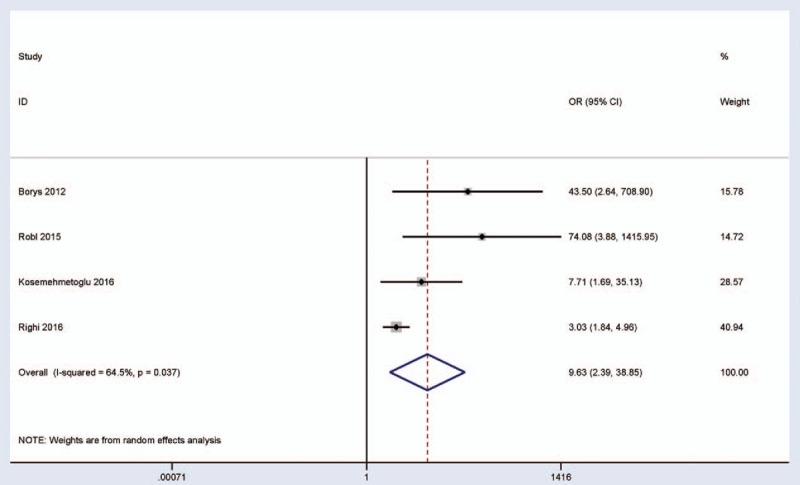
Forest plot for the association between P16 protein expression and pathologic complete response using multivariate logistic regression analysis.
3.4. Survival analysis of P16 protein expression on high-grade osteosarcoma patients
There were 2 studies estimating the association between P16 expression and OS of 404 patients with high-grade osteosarcoma in multivariate regression analysis (Fig. 5). The pooled HR for OS showed that the expression of P16 protein was not correlated with the prognosis of high-grade osteosarcoma patients in OS (HR = 0.33, 95% CI = 0.02–5.74, P = .448). Only one study involving 357 patients with high-grade osteosarcoma reported that no significant relationship was observed between P16 expression and DFS for multivariate analysis (HR = 1.23, 95% CI = 0.86–1.76, P = .263).[23]
Figure 5.
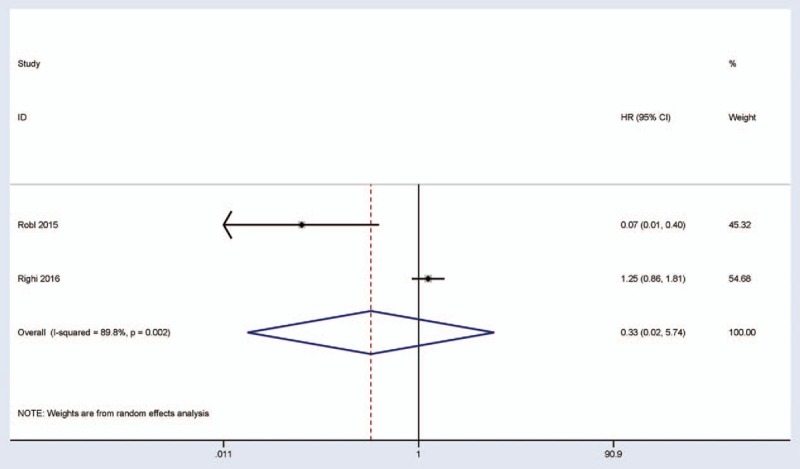
Forest plot for the association between P16 expression and overall survival by multivariate regression analysis.
3.5. Sensitivity analysis
A substantial heterogeneity was found between p16 expression and pathologic complete response after neoadjuvant chemotherapy using multivariate analysis (PY=Y.037Y<Y.1). When we removed this study (Robl et al, 2015),[30] and recalculated the overall OR (OR = 3.55, 95% CI = 2.23–5.65, P < .001), with no significant evidence of heterogeneity (P = .106). The analysis indicated the stability of our result.
4. Discussion
Patients with osteosarcoma with local relapse and distant metastasis have a relatively high mortality.[32] The conventional therapy of high-grade osteosarcoma consists of neoadjuvant chemotherapy and postoperative adjuvant chemotherapy.[33] The assessment of the histological response to chemotherapy is considered to be a good predictor of survival in osteosarcoma, and a good pathologic response is ≥90% tumor necrosis.[34] Some molecular markers such as STAT3 and ERK1 expressions are noted to be correlated with poor response to chemotherapy.[35] The p16 tumor suppressor protein has been reported in several types of human cancers, and it is suggested as a potential predictor in the progression and prognosis of cancer.[36,37] The P16 protein was frequently expressed in high-grade osteosarcoma in this study, with a varying frequency of 36.2%[30] to 70.6%.[23] Therefore, the purpose of this study is to investigate whether P16 protein expression using IHC can be used as a predictive biomarker of response to neoadjuvant chemotherapy and prognostic marker in 527 patients with high-grade osteosarcoma.
We analyzed the relationship between the expression of P16 protein and clinicopathological characteristics in high-grade osteosarcoma. The results suggested that significant correlation was not found between P16 expression and sex, tumor histotypes, or location of high-grade osteosarcoma patients (P > .1).
A study with 47 patients with high-grade osteosarcoma after neoadjuvant chemotherapy suggests that P16 protein expression is related to OS in multivariate regression analysis.[30] Righi et al[23] reported that the expression of P16 protein using multivariate analysis was not found to be correlated with OS and DFS in 357 high-grade osteosarcoma patients receiving neoadjuvant chemotherapy. These 2 studies were pooled, and the present result suggested that P16 protein expression in a larger research of 404 patients with high-grade osteosarcoma was not associated with the prognosis of patients in OS.
We also evaluated the association between P16 protein expression and neoadjuvant chemotherapy response. Our findings indicated that P16 protein expression was correlated with “favorable” neoadjuvant chemotherapy in P16-expressing tumors versus P16-nonexpressing tumors (OR = 4.69, P < .001). Furthermore, the initial results involving 527 whites with high-grade osteosarcoma using multivariate analysis were extracted, the pooled OR was 9.63 (95% CI = 2.39–38.85, P = .001), which suggested that the expression of P16 protein might be used as a promising noninvasive marker to predict the postchemotherapy necrotic response in the white population with high-grade osteosarcoma.
Several limitations should be carefully considered in the present study. First, our results included 527 patients with high-grade osteosarcoma, the total sample sizes were <1000,[38] which may lack statistically vigorous power. Second, only eligible studies published in English were included in our meta-analysis. Articles published in other languages other than English were excluded because of unreadable contents, and conference abstracts were also excluded based on insufficient information. Third, only white population were included; additional large-scale studies with larger sample size are needed in the future, particularly in the Asian and African populations.
In summary, our findings suggested that among high-grade osteosarcoma patients, the use of P16 protein expression as a noninvasive biomarker may be a potential predictor of the response to neoadjuvant chemotherapy in clinical application. However, it was not associated with the prognosis of patients in OS and DFS in multivariate analysis. Additional clinical researches with larger cases should be essential to further validate the value of P16 expression in Asians and Africans.
Footnotes
Abbreviations: 95% CI = 95% confidence interval, CDKN2A = cyclin-dependent kinase inhibitor 2A, DFS = disease-free survival, HR = hazard ratio, IHC = immunohistochemistry, OR = odds ratio, OS = overall survival.
Authors’ Contributions: Y.T. and Q.P. contributed to the conception and design of this study; Y.T., C.Y., Z.G., Y.F., X.Y., B.L., H.Z., J.W., and W.L. contributed to the retrieval of articles, the extraction of data, the calculation of data, and the design of the figures and tables; all the authors approved the final article.
This research was supported by grants from Natural Science Foundation of Ningbo, China (No. 2014A610254), Key Program of Clinical Specialty Disciplines of Ningbo, China (No. 2013–88), Key Program of Medical Disciplines of Ningbo, China (No. 2016–55) and Program of Medical Science and Technology of Ningbo, China (No. 2016A49).
The authors report no conflicts of interest.
References
- [1].Fox MG, Trotta BM. Osteosarcoma: review of the various types with emphasis on recent advancements in imaging. Semin Musculoskelet Radiol 2013;17:123–36. [DOI] [PubMed] [Google Scholar]
- [2].Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res 2009;152:3–13. [DOI] [PubMed] [Google Scholar]
- [3].Picci P. Osteosarcoma (osteogenic sarcoma). Orphanet J Rare Dis 2007;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jeon DG, Song WS. How can survival be improved in localized osteosarcoma? Expert Rev Anticancer Ther 2010;10:1313–25. [DOI] [PubMed] [Google Scholar]
- [5].Arndt CA, Crist WM. Common musculoskeletal tumors of childhood and adolescence. N Engl J Med 1999;341:342–52. [DOI] [PubMed] [Google Scholar]
- [6].Hameed M, Dorfman H. Primary malignant bone tumors–recent developments. Semin Diagnost Pathol 2011;28:86–101. [DOI] [PubMed] [Google Scholar]
- [7].Carty CP, Dickinson IC, Watts MC, et al. Impairment and disability following limb salvage procedures for bone sarcoma. Knee 2009;16:405–8. [DOI] [PubMed] [Google Scholar]
- [8].Meyers PA, Schwartz CL, Krailo M, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol 2005;23:2004–11. [DOI] [PubMed] [Google Scholar]
- [9].Goorin AM, Schwartzentruber DJ, Devidas M, et al. Presurgical chemotherapy compared with immediate surgery and adjuvant chemotherapy for nonmetastatic osteosarcoma: Pediatric Oncology Group Study POG-8651. J Clin Oncol 2003;21:1574–80. [DOI] [PubMed] [Google Scholar]
- [10].Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009;115:1531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatr Drugs 2008;10:315–27. [DOI] [PubMed] [Google Scholar]
- [12].Jones KB, Salah Z, Del Mare S, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res 2012;72:1865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bjornland K, Flatmark K, Pettersen S, et al. Matrix metalloproteinases participate in osteosarcoma invasion. J Surg Res 2005;127:151–6. [DOI] [PubMed] [Google Scholar]
- [14].Pakos EE, Kyzas PA, Ioannidis JP. Prognostic significance of TP53 tumor suppressor gene expression and mutations in human osteosarcoma: a meta-analysis. Clin Cancer Res 2004;10(18 pt 1):6208–14. [DOI] [PubMed] [Google Scholar]
- [15].Entz-Werle N, Schneider A, Kalifa C, et al. Genetic alterations in primary osteosarcoma from 54 children and adolescents by targeted allelotyping. Br J Cancer 2003;88:1925–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Agarwal P, Sandey M, DeInnocentes P, et al. Tumor suppressor gene p16/INK4A/CDKN2A-dependent regulation into and out of the cell cycle in a spontaneous canine model of breast cancer. J Cell Biochem 2013;114:1355–63. [DOI] [PubMed] [Google Scholar]
- [17].Lukas J, Parry D, Aagaard L, et al. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 1995;375:503–6. [DOI] [PubMed] [Google Scholar]
- [18].Mitra AP, Hansel DE, Cote RJ. Prognostic value of cell-cycle regulation biomarkers in bladder cancer. Semin Oncol 2012;39:524–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Romagosa C, Simonetti S, Lopez-Vicente L, et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 2011;30:2087–97. [DOI] [PubMed] [Google Scholar]
- [20].Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res 2001;264:42–55. [DOI] [PubMed] [Google Scholar]
- [21].Mohseny AB, Szuhai K, Romeo S, et al. Osteosarcoma originates from mesenchymal stem cells in consequence of aneuploidization and genomic loss of Cdkn2. J Pathol 2009;219:294–305. [DOI] [PubMed] [Google Scholar]
- [22].Park YB, Park MJ, Kimura K, et al. Alterations in the INK4a/ARF locus and their effects on the growth of human osteosarcoma cell lines. Cancer Genet Cytogenet 2002;133:105–11. [DOI] [PubMed] [Google Scholar]
- [23].Righi A, Gambarotti M, Sbaraglia M, et al. P16 expression as a prognostic and predictive marker in high grade localized osteosarcoma of the extremities: an analysis of 357 cases. Human Pathol 2016;58:15–23. [DOI] [PubMed] [Google Scholar]
- [24].Bu J, Li H, Liu LH, et al. P16INK4a overexpression and survival in osteosarcoma patients: a meta analysis. Int J Clin Exp Pathol 2014;7:6091–6. [PMC free article] [PubMed] [Google Scholar]
- [25].Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics 2005;21:3672–3. [DOI] [PubMed] [Google Scholar]
- [26].Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med 1997;127:820–6. [DOI] [PubMed] [Google Scholar]
- [27].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. Bmj 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med 1996;15:1237–48. [DOI] [PubMed] [Google Scholar]
- [29].Kosemehmetoglu K, Ardic F, Karslioglu Y, et al. p16 expression predicts neoadjuvant tumor necrosis in osteosarcomas: reappraisal with a larger series using whole sections. Hum Pathol 2016;50:170–5. [DOI] [PubMed] [Google Scholar]
- [30].Robl B, Pauli C, Botter SM, et al. Prognostic value of tumor suppressors in osteosarcoma before and after neoadjuvant chemotherapy. BMC Cancer 2015;15:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Borys D, Canter RJ, Hoch B, et al. P16 expression predicts necrotic response among patients with osteosarcoma receiving neoadjuvant chemotherapy. Hum Pathol 2012;43:1948–54. [DOI] [PubMed] [Google Scholar]
- [32].Ta HT, Dass CR, Choong PF, et al. Osteosarcoma treatment: state of the art. Cancer metastasis Rev 2009;28:247–63. [DOI] [PubMed] [Google Scholar]
- [33].Chui MH, Kandel RA, Wong M, et al. Histopathologic features of prognostic significance in high-grade osteosarcoma. Arch Pathol Lab Med 2016. [DOI] [PubMed] [Google Scholar]
- [34].Bacci G, Mercuri M, Longhi A, et al. Grade of chemotherapy-induced necrosis as a predictor of local and systemic control in 881 patients with non-metastatic osteosarcoma of the extremities treated with neoadjuvant chemotherapy in a single institution. Eur J Cancer 2005;41:2079–85. [DOI] [PubMed] [Google Scholar]
- [35].Salas S, Jiguet-Jiglaire C, Campion L, et al. Correlation between ERK1 and STAT3 expression and chemoresistance in patients with conventional osteosarcoma. BMC Cancer 2014;14:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Karim RZ, Gerega SK, Yang YH, et al. p16 and pRb immunohistochemical expression increases with increasing tumour grade in mammary phyllodes tumours. Histopathology 2010;56:868–75. [DOI] [PubMed] [Google Scholar]
- [37].Maitra A, Roberts H, Weinberg AG, et al. Loss of p16(INK4a) expression correlates with decreased survival in pediatric osteosarcomas. Int J Cancer 2001;95:34–8. [DOI] [PubMed] [Google Scholar]
- [38].Karahalios A, Baglietto L, Carlin JB, et al. A review of the reporting and handling of missing data in cohort studies with repeated assessment of exposure measures. BMC Med Res Methodol 2012;12:96. [DOI] [PMC free article] [PubMed] [Google Scholar]


