Abstract
In the most studied rhizobium-legume interactions, the host plant supplies the symbiont with homocitrate, an essential co-factor of the nitrogenase enzyme complex, via the expression of a nodule-specific homocitrate synthase FEN1. Photosynthetic bradyrhizobia interacting with Nod factor (NF) dependent and NF-independent Aeschynomene legumes are able to synthesize homocitrate themselves as they contain a nifV gene encoding a homocitrate synthase. Here, we show that in the model strain ORS285, nifV is required for free-living and symbiotic dinitrogen fixation with NF-independent Aeschynomene species. In contrast, in symbiosis with NF-dependent Aeschynomene species, the nifV requirement for efficient nitrogen fixation was found to be host plant dependent. Interestingly, orthologs of FEN1 were found in both NF-dependent and NF-independent Aeschynomene species. However, a high nodule specific induction of FEN1 expression was only observed in A. afraspera, a host plant in which nifV is not required for symbiotic dinitrogen fixation. These data indicate that efficient symbiotic nitrogen fixation in many of the tested Aeschynomene species requires rhizobial homocitrate synthesis. Considering that more than 10% of the fully sequenced rhizobium strains do contain a nifV gene, the Aeschynomene/photosynthetic Bradyrhizobium interaction is likely not the only rhizobium/legume symbiosis where rhizobial nifV expression is required.
Introduction
Nitrogen is an essential element for all living organisms. On earth the major source of nitrogen is atmospheric dinitrogen, which is fixed by microorganism (diazotrophs) that are able to reduce dinitrogen to ammonium by a nitrogenase enzyme complex. Leguminous plants establish a nitrogen-fixing symbiotic interaction with soil bacteria commonly called rhizobia. This symbiosis is a major contributor to the global nitrogen cycle and enables the host legumes to grow without an exogenous nitrogen source. In general, rhizobia induce the formation of a new organ, the nodule, on the roots of their host plant. The plant cells in the nodule are colonized intracellularly by the rhizobia which differentiate into an endosymbiotic form, the bacteroids, able to reduce atmospheric dinitrogen to the benefit of the plant. In turn, the plant supplies the bacteroids with the required carbon sources.
For nitrogen fixation, rhizobia use a molybdenum (Mo)-nitrogenase (EC 1.18.2.1). The enzymatic complex is composed of two components: the Fe protein and the MoFe protein containing a P-cluster and iron-molybdenum cofactor (FeMo-co). In the well-studied free-living diazotroph, Klebsiella pneumonia, the formation of an active nitrogenase enzyme complex depends on a cluster of 20 (nif) genes (for review see: ref. 1). The structural genes for the Fe protein and α- and β-subunits of the MoFe protein are encoded by the nifH, nifD and nifK genes, respectively. However, the synthesis of these structural proteins is not sufficient to obtain an active nitrogenase complex. It requires additional nif genes which play a role in the biosynthesis and the assembly of the FeMo-cofactor, in electron transport and in nitrogenase regulation (see for review: ref. 2). In contrast to free-living diazotrophs, the number of nif genes in rhizobia is very variable (for review see: ref. 3). Remarkably, the majority of the rhizobia lack the nifV gene which encodes a homocitrate synthase that catalyzes the condensation of acetyl coenzyme A and 2-oxoglutarate. Homocitrate is a component of the FeMo-cofactor present in the catalytic center of dinitrogenase that is absolutely required for a proper functioning of the nitrogenase enzyme complex4 which raises the question how the rhizobia are able to fix nitrogen during symbiosis. Recently, it has been demonstrated that Lotus japonicus expresses a nodule specific homocitrate synthase (FEN1) that compensates for the absence of homocitrate synthase activity in the bacterial partner Mesorhizobium loti 5. Azorhizobium caulinodans ORS571 and photosynthetic Bradyrhizobium strains are examples of rhizobia that contain a nifV gene. However, as these rhizobia are capable to fix dinitrogen under free-living conditions6, 7, we can ask if the presence of the nifV gene in these rhizobia is related to this capacity or because their host plant are unable to supply homocitrate during symbiosis.
To investigate this question, we have constructed a nifV deletion mutant in the photosynthetic Bradyrhizobium strain ORS285. We preferentially selected this strain as it can establish a nitrogen symbiosis with two distinct groups of Aeschynomene species that are discriminated by their use of a Nod-Factor (NF) dependent or a NF-independent mechanism to establish a symbiotic interaction. We correlated the nodulation phenotypes with the presence and expression of FEN1 orthologs in A. afraspera and A. evenia, representatives of the NF-dependent and NF-independent Aeschynomene groups, respectively.
Results
Deletion of nifV results in a reduced nitrogenase enzyme activity in free-living conditions
Genomic analysis revealed the presence of one nifV gene in the ORS285 strain. This gene is localized in a chromosomic region that contains other nif genes (nifH/Q/P/W) and genes that have been shown to be necessary for an efficient nitrogen fixation (fixABCX, mopBmodCD) (Fig. S1). To analyze the importance of nifV in nitrogen fixation, we have constructed a nifV deletion mutant by double crossing over and analyzed its nitrogenase activity under free-living conditions using the acetylene reduction assay (ARA). Under growth conditions that induce nitrogenase genes, i.e. under low oxygen tension and absence of a combined source of nitrogen, the kinetics of ethylene formation by the ΔnifV mutant was drastically reduced as compared to the WT strain (Fig. 1A). Interestingly, analysis of the chromatograms revealed that gas samples from the ΔnifV mutant contained an additional volatile molecule eluting just before ethylene (Fig. 1B). A similar volatile molecule was observed in gaschromatograms from nifV mutants of other diazotrophs and identified as ethane8, 9. Therefore, the additional peak in the gaschromatogram we have indicated as “ethane” in the rest of the text. The addition of homocitrate to the growth medium or the re-introduction of a plasmid containing the nifV gene restores completely the nitrogenase activity of the ΔnifV mutant (Fig. 1C; Fig. S2A and B). This indicates that the observed effects in the ΔnifV mutant are solely due to the absence of the homocitrate synthase NifV and not to pleiotropic effects of the nifV deletion on the expression of downstream genes (cysE/nifW).
Figure 1.
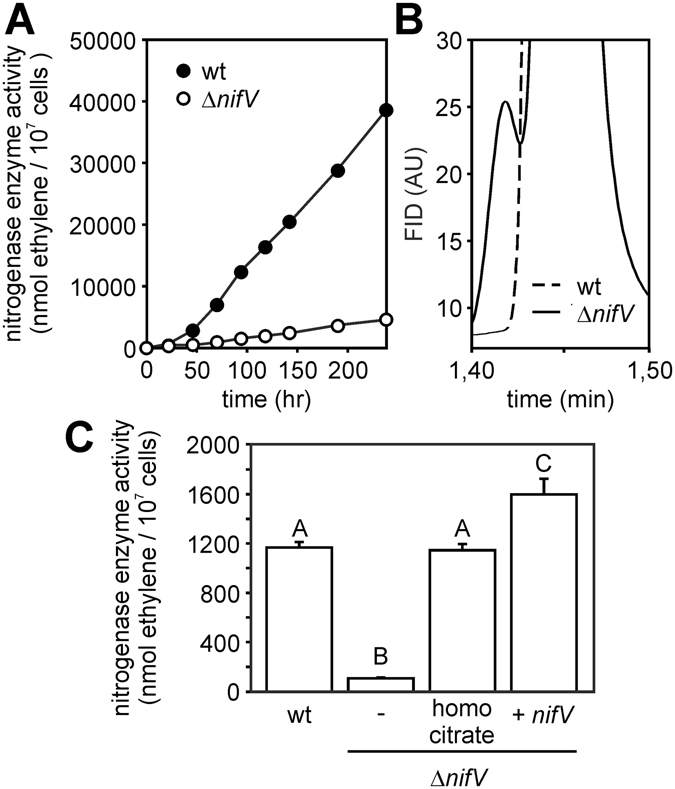
ORS285 ΔnifV mutant displays low in vitro acetylene reducing activity which is restored by homocitrate addition or re-introducing a complete nifV gene. (A) Ethylene production by ORS285 and ORS285 ΔnifV cultures grown in 150 ml vials containing BNM-B medium and 10% acetylene gas at different times post-inoculation. (B) Chromatogram of gas-samples taken from ORS285 (wt) and ORS285 ΔnifV cultures 8 days post-inoculation. WT: dashed line. ΔnifV mutant: solid line. (C) Ethylene production of ORS285 and derivatives grown for 7 days in vacuette® tubes containing BNM-B medium and 10% acetylene gas in the absence or presence of 10 mM homocitrate. The mean amount of produced ethylene per tube (n = 3) is indicated. Error bars represent standard errors of the mean. An ANOVA was performed among the different conditions, followed by Tukey’s post hoc analysis (P < 0.05). The different letters indicate groups that differ significantly.
Rhizobial homocitrate synthase is required for an efficient symbiotic interaction with NF-independent Aeschynomene species
Dependent on the host plant, Bradyrhizobium ORS285 uses a NF-dependent or a NF-independent mechanism to establish a symbiotic interaction10. To investigate whether the absence of nifV affects the NF-independent interaction, we infected A. evenia with both the WT and the ΔnifV mutant. Observations done at 22 days post infection (dpi) showed drastic difference between the plants, the ΔnifV infected plants had typical nitrogen starvation symptoms such as foliar chlorosis and reduced plant growth as observed for the non-inoculated plant (Fig. 2A). Furthermore, while the plants infected with the WT strain harbor only red/pink colored nodules, nodules elicited by the ΔnifV mutant were heterogeneous in color and pink, yellow and green nodules were observed (Fig. 2A and B). The green color is indicative for leghaemoglobin degradation. Analyzing the kinetics of nodule formation showed that after 10 dpi the number of nodules on plants infected by the ΔnifV strain continued to increase whereas in plants infected by the WT strain the number of nodules started to stabilize. As a result, at 22 dpi, the number of nodules on ΔnifV infected plants was approximately twice the number found on plants inoculated with the WT strain (Fig. 2C). This increase in the nodule number is similar to what has been observed upon infection with nitrogenase minus mutants11, and could be attributed to a phenomenon termed autoregulation of nodulation that ensures a balance between nodule formation and energy requirements in legumes12. The acetylene reduction assay (ARA) showed that nodules of plants inoculated with the ΔnifV mutant strain had a very low nitrogenase enzyme activity (~10% of WT nodules; Fig. 2D) and produced “ethane” (Fig. S3A). All the observed phenotypes were absent when A. evenia plants were infected with an ORS285 ΔnifV strain that contained a plasmid carrying the nifV gene (Fig. 2A–D). Together, these data indicate that the nifV gene plays a critical role in the nitrogenase activity of the ORS285 strain during the symbiosis with A. evenia. The Aeschynomene species that form a NF-independent symbiotic interaction fall into a single clade13. To investigate whether the dependence on the rhizobial nifV gene is a generality within this clade, we infected nine other NF-independent species with the ORS285 ΔnifV mutant strain. With all tested species, plants inoculated with the ΔnifV mutant strain showed nitrogen starvation symptoms (Table 1). We must remark that for 3 species (A. virginica, A. pratensis, A. selloi) there was not a significant difference in the nitrogenase enzyme activity as measured by the ARA assay between plants inoculated with the WT and ΔnifV mutant strain. However, the formation of “ethane” in the ARA assay, an increased number of nodules, the presence of nodules with signs of senescence and the reduced stimulation of plant growth evidenced a nitrogenase enzyme complex that reduces dinitrogen inefficiently (Table 1). Taken together, these data indicate that the presence of the rhizobial homocitrate synthase NifV is required for an efficient symbiotic interaction with all tested NF-independent Aeschynomene species.
Figure 2.
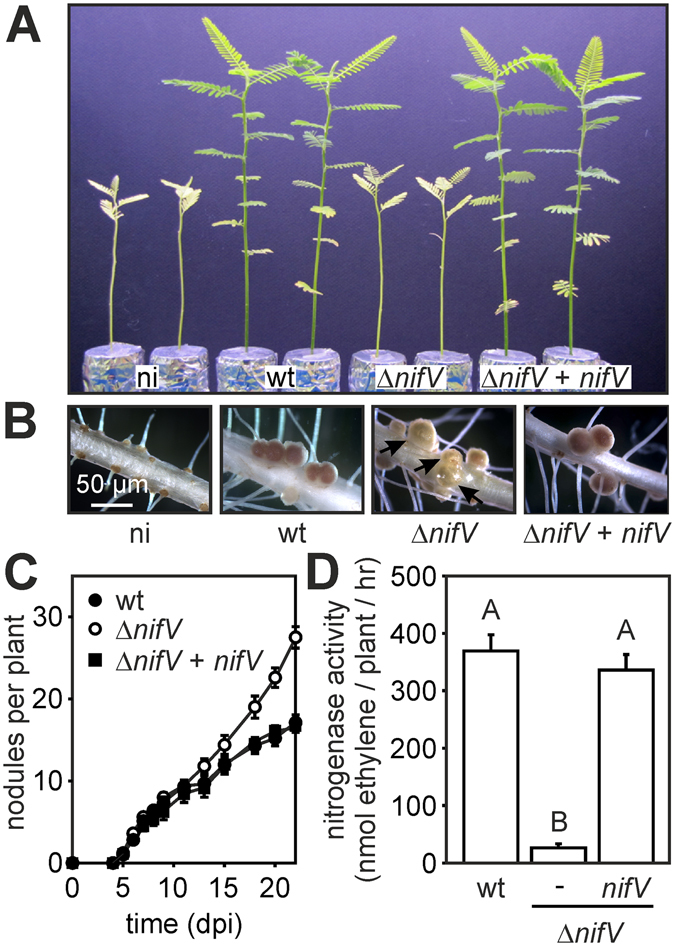
ORS285 nifV deletion affects the symbiotic interaction with Aeschynomene evenia (CIAT22838). (A) Comparison of the growth of A. evenia plants inoculated with ORS285, ORS285 ΔnifV and ORS285 ΔnifV+pMG105-nifV. Non-inoculated plants (ni) were used as control. (B) Mature nodules on A. evenia plants inoculated with different ORS285 derivatives. Note the presence of green colored nodules (indicated with a black arrow) on A. evenia plants inoculated with the ORS285 ΔnifV strain. (C) Nodulation kinetics of Bradyrhizobium ORS285 (wt), ΔnifV and ΔnifV+pMG105-nifV (ΔnifV+nifV) derivatives on A. evenia plants. The mean number of nodules per plant (n = 10) at various days post infection (dpi) is presented. Error bars represent standard errors of the mean. (D) Acetylene reducing activity of A. evenia plants inoculated with Bradyrhizobium ORS285, ΔnifV and ΔnifV+nifV derivatives at 22 dpi. The mean amount of produced ethylene per hour and per plant is indicated. Error bars represent standard errors of the mean (n = 10). An ANOVA was performed among the different conditions, followed by Tukey’s post hoc analysis (P < 0.05). The different letters above the bars indicate groups that differ significantly.
Table 1.
Characteristics of the symbiotic interaction between different NF-independent Aeschynomene species and the WT and ΔnifV mutant of Bradyrhizobium ORS285.
| Plant species | Growth stimulation plants | Mean number of nodules/plant | Legheamoglobin degradation (green nodules) | Nitrogenase enzyme activity (nmol ethylene/hr/plant) | “Ethane formation” | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | ΔnifV | WT | ΔnifV | WT | ΔnifV | WT | ΔnifV | WT | ΔnifV | |
| A. evenia | +++ | − | 17 ± 1 (A) | 28 ± 1 (B) | no | yes | 370 ± 28 (A) | 27 ± 7 (B) | no | yes |
| A. indica | +++ | − | 33 ± 2 (A) | 49 ± 3 (B) | no | yes | 590 ± 26 (A) | 340 ± 120 (B) | no | yes |
| A. scabra | +++ | − | 17 ± 1 (A) | 33 ± 1 (B) | no | yes | 534 ± 89 (A) | 190 ± 17 (B) | no | yes |
| A. sensitiva | +++ | − | 14 ± 3 (A) | 27 ± 3 (B) | no | yes | 332 ± 34 (A) | 69 ± 24 (B) | no | yes |
| A. deamii | +++ | − | 24 ± 1 (A) | 45 ± 2 (B) | no | yes | 665 ± 59 (A) | 31 ± 10 (B) | no | yes |
| A. denticulata | +++ | − | 18 ± 1 (A) | 48 ± 2 (B) | no | yes | 402 ± 29 (A) | 227 ± 30 (B) | no | yes |
| A. virginica | +++ | + | 39 ± 2 (A) | 82 ± 3(B) | no | yes | 867 ± 89 (A) | 819 ± 35 (A) | no | yes |
| A. tambacoudensis | ++ | − | 25 ± 3 (A) | 49 ± 1(B) | no | yes | 247 ± 13 (A) | 80 ± 13 (B) | no | yes |
| A. pratensis | +++ | + | 11 ± 1 (A) | 23 ± 1 (B) | no | yes | 202 ± 62 (A) | 151 ± 12 (A) | no | yes |
| A. selloi | +++ | + | 20 ± 2 (A) | 29 ± 1 (B) | no | yes | 574 ± 55 (A) | 482 ± 34 (A) | no | yes |
Seeds of different Aeschynomene species were sterilized, germinated and seedlings were inoculated with Bradyrhizobium ORS285 and Bradyrhizobium ORS285 ΔnifV, respectively. At 22 (A. evenia) or 28 dpi (others), the plant growth was compared with non–inoculated control plants and the mean nodule number, mature nodule phenotype and mean nitrogenase enzyme activity as analysed by the ARA assay was determined. +++: no N-starvation signs and plants are much better developed than the non-inoculated control plants; ++: no N-starvation signs and plants are better developed than the non-inoculated control plants; +: plants are better developed than the non-inoculated control plants but have signs of foliar chlorosis; −: no difference with non-inoculated control plants. ±Indicates the standard error of the mean. Letters in brackets in the table represent conditions with significant difference according to the Tukey’s test (P < 0.05).
In NF-dependent Aeschynomene species the requirement for rhizobial NifV depends on the host plant
To investigate the role of NifV in the NF-dependent symbiotic interaction, we infected A. afraspera plants with the ORS285 ΔnifV mutant. At 22 dpi, plants inoculated with the ΔnifV mutant were in all phenotypic aspects (growth, nodule number and color) indistinguishable from plants inoculated with the WT strain (Fig. 3A,B and C). Also no “ethane” formation was detected in the ARA assay (Fig. S3B) and the kinetics of nodule formation and nitrogenase activity were similar as observed with the WT strain (Fig. 3C and D). In the group of NF-dependent Aeschynomene species, only few can form efficient nitrogen fixing nodules with the ORS285 strain13. A. nilotica plants that belong to this small group, have typical nitrogen starvation symptoms and all other phenotypes as observed with the group of NF-independent Aeschynomene species when inoculated with the ΔnifV mutant (Fig. 4A–D,B; Fig. S3C). This indicates that in contrast to A. afraspera, A. nilotica plants are unable to compensate for the absence of NifV in ORS285. Thus, in the group of NF-dependent Aeschynomene species, the requirement of NifV for a symbiotic nitrogenase activity differs according to the host plant.
Figure 3.
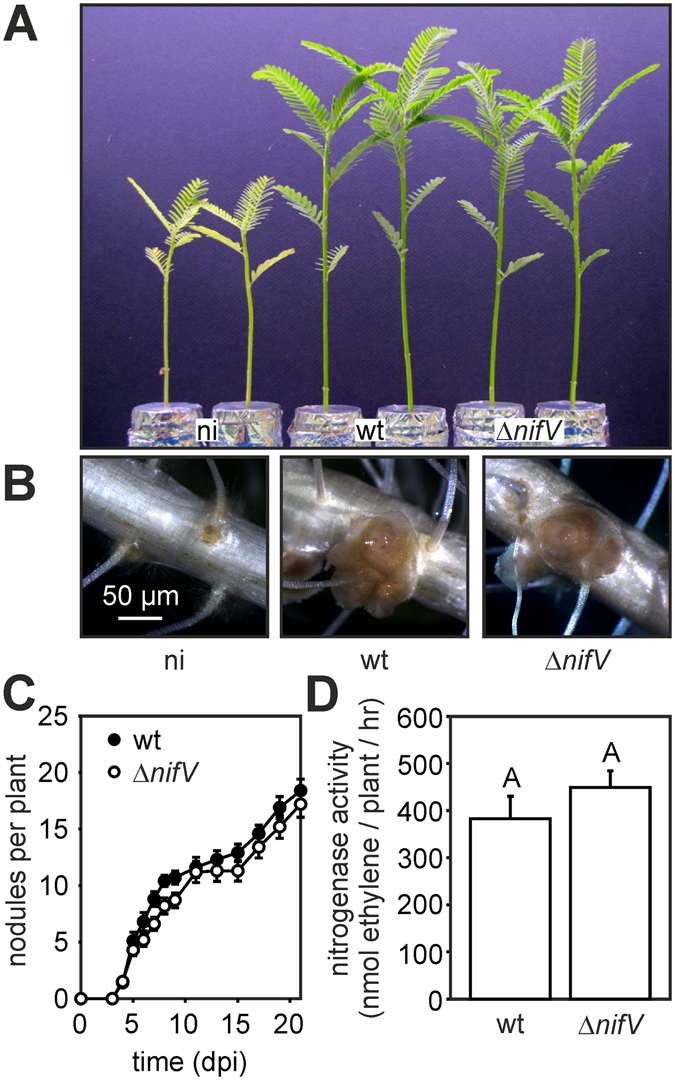
ORS285 nifV deletion does not affect the symbiotic interaction with Aeschynomene afraspera (LSTM #1). (A) Comparison of the growth of A. afraspera plants inoculated with ORS285 (wt) and ORS285 ΔnifV. Non-inoculated plants (ni) were used as control. (B) Mature nodules on A. afraspera plants inoculated with ORS285 and ORS285 ΔnifV. (C) Nodulation kinetics of Bradyrhizobium ORS285 (wt) and ORS285 ΔnifV (∆nifV) on A. afraspera plants. The mean number of nodules per plant (n = 10) at various days post infection (dpi) is presented. (D) Acetylene reducing activity of A. afraspera plants inoculated with Bradyrhizobium ORS285 (wt) and ORS285 ΔnifV at 21 dpi. The mean amount of produced ethylene per hour and per plant is indicated. Error bars represent standard errors of the mean (n = 10). Tukey’s post hoc analysis (P < 0.05) showed no significant difference in acetylene reduction between plants inoculated with the two strains.
Figure 4.
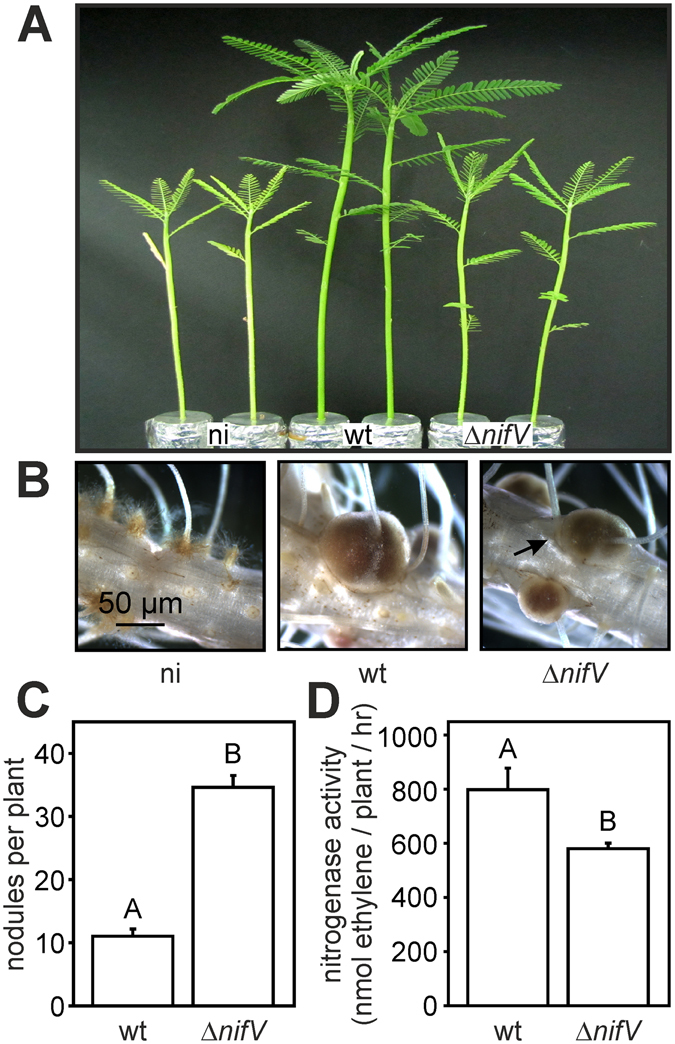
ORS285 nifV deletion affects the symbiotic interaction with Aeschynomene nilotica (IRRI 014040). (A) Comparison of the growth of A. nilotica plants inoculated with ORS285 and ORS285 ΔnifV at 28 days post infection (dpi). Non-inoculated plants (ni) were used as control. (B) Mature nodules on A. nilotica plants inoculated with ORS285 and ORS285 ΔnifV. Note the presence of green colored nodules (black arrow) on A. nilotica plants inoculated with the ORS285 ΔnifV strain. (C) Number of root nodules on A. nilotica plants inoculated with Bradyrhizobium ORS285 and Bradyrhizobium ORS285 ΔnifV, respectively. The mean number of nodules per plant (n = 5) at 28 dpi is presented. (D) Acetylene reducing activity of A. nilotica plants inoculated with Bradyrhizobium ORS285 and ORS285 ΔnifV at 28 dpi. The mean amount of produced ethylene per hour and per plant (n = 5) is indicated. In (C) and (D) error bars represent standard errors of the mean and letters represent conditions with significant difference according to the Tukey’s test (P < 0.05).
Aeschynomene evenia and Aeschynomene afraspera contain putative orthologs of Lotus japonicus FEN1
L. japonicus and G. max express a nodule-specific homocitrate synthase gene annotated respectively FEN1 and GmN565, 14. Interestingly, previous analyses of the transcriptomes of A. evenia (CIAT22838) that included nodule tissue failed to identify a FEN1 ortholog15. In order to analyze the presence and expression of FEN1-like genes in different Aeschynomene species, we made use of genomic data obtained in the frame of an ongoing genome sequencing project for A. evenia (CIAT22838) to perform a blast search using L. japonicus FEN1 as query. This resulted in one gene-product that after phylogenetic analysis clustered in a clade with FEN1 of L. japonicus (Fig. 5A). By PCR, cloning, sequencing and transriptomic data analysis (see material and method section SI) using this A. evenia (CIAT22838) FEN1 sequence as basis, we obtained four different sequences of putative FEN1 genes for A. afraspera and one sequence for another A. evenia line, PI 225551. This difference in the number of FEN1 homologs between these two species is not surprising considering that A. evenia is diploid whereas A. afraspera is octoploid16. Phylogenetic analysis showed that all the gene products clustered in a clade containing L. japonicus FEN1 (Fig. 5A). This suggests that FEN1 is present in a single copy in the 2x A. evenia genome and that 4 paralogs are present in the 8x A. afraspera genome.
Figure 5.
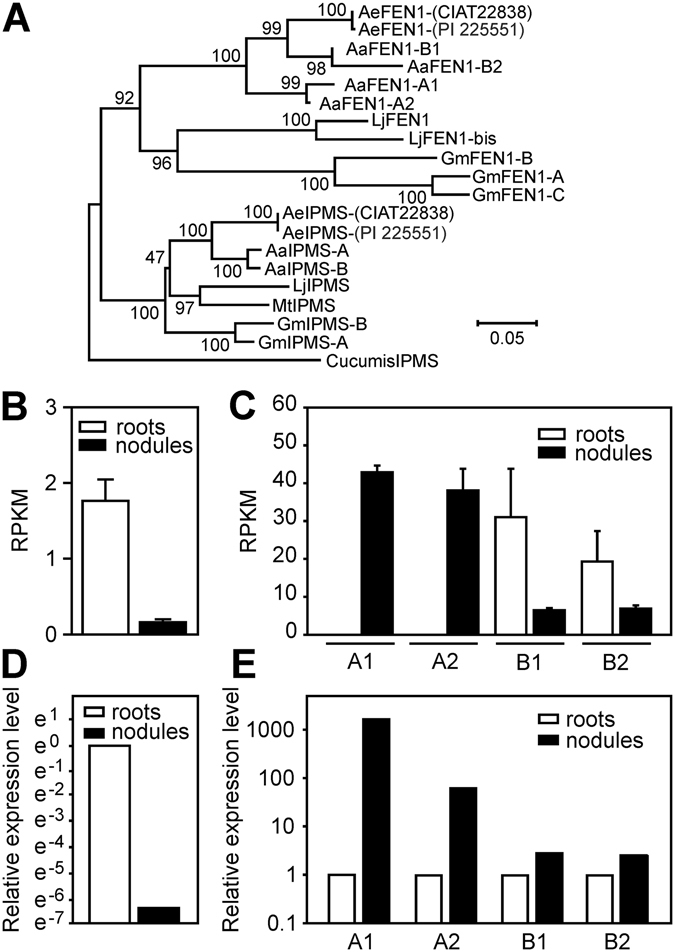
The Aeschynomene legumes A. evenia and A. afraspera contain FEN1 homoloques but only nodule specific expression is observed in A. afraspera plants. (A) Phylogenie based on FEN1 and isopropylmalate synthase (IPMS) sequences obtained from genomic and transcriptome databases of A. evenia (CIAT22838/PI 225551) and A. afraspera (LSTM #1). Cuccumis, Glycine max, Lotus japonicus and Medicago truncatula IPMS and FEN1 sequences were obtained from Genebank. -A, -A1, -B, -B1, bis and -C indicate different copies found in (polyploid) species. Cuccumis IPMS was used as outgroup. Numbers at nodes represent bootstrap values (% of 1000 replicates). (B) Transcript abundance reads (reads per kilobase per million; RPKM) of FEN1 in root and nodule tissue of A. evenia (PI 225551). (C) Transcript abundance reads (RPKM) of the different FEN1 homologues in root and nodule tissue of A. afraspera (LSTM #1) Error bars as shown in (B) and (C) indicate standard errors of the means of three biological replicates. (D) Relative FEN1 expression level in A. evenia (PI 225551) nodules elicited by Bradyrhizobium ORS285 at 8 dpi. (E) Relative expression level of the different FEN1 homoloques in A. afraspera (LSTM #1) nodules elicited by Bradyrhizobium ORS285 at 8 dpi. The relative expression level of the different FEN1 homologues as shown in (D), (E) was determined by RT-qPCR and normalized by the expression of Elongation Factor 1α. Non-inoculated roots were used as control.
Analyzing transcriptomic data showed that in A. evenia (PI 225551) the expression level of the identified FEN1 gene is low and not nodule-specific (Fig. 5B), in accordance with the reported absence of expression for the other A. evenia line CIAT2283815. However, in A. afraspera two of the four FEN1 copies (A1 and A2) are highly expressed specifically in nodules during symbiosis with Bradyrhizobium ORS285 (Fig. 5C). RT-Q-PCR analysis using specific primers for the individual FEN1 copies confirmed the experimental data as obtained by RNA-seq analysis (Fig. 5D and E). Hence, for these two tested Aeschynomene species, the absence of FEN1 expression correlates with the NifV requirement for efficient nitrogen fixation in nodules.
Discussion
How do rhizobia fix nitrogen during symbiosis while they lack the nifV gene that is required for homocitrate synthesis, an essential co-factor of the nitrogenase enzyme complex? This important issue has been resolved in the case of the L. japonicus – Mesorhizobium loti symbiosis for which it has been shown that the plant overcomes the lack of nifV in the bacterial partner via a nodule specific expression of an homocitrate synthase homolog, FEN15. However, the question remains if this plant homocitrate supplementation constitutes a general paradigm for all the rhizobium/legume symbioses. This question is particularly meaningful in the case of symbiotic interactions involving rhizobia that do contain a nifV gene, such as photosynthetic bradyrhizobia and A. caulinodans.
Here, by studying the symbiotic interaction of a nifV mutant of the photosynthetic Bradyrhizobium strain ORS285 and different Aeschynomene species, we demonstrate that not all legumeous plants supply the rhizobial partner with (sufficient) homocitrate for nitrogen fixation. In particular for all the NF-independent Aeschynomene species tested, we observed an important effect of the ΔnifV mutation on the symbiotic efficiency indicating the inability of these plant species to compensate for the absence of nifV (Fig. 2 and Table 1). It is to note that for 3 out of the 9 species tested, the symptoms of the ΔnifV mutation are not as drastic as observed for A. evenia (Table 1). In these cases the measured nitrogenase enzyme activity of the ΔnifV inoculated plants was close to these of the WT strain and a weak benefit on the plant growth by inoculation with the ΔnifV mutant was observed. The nitrogenase enzyme activity as measured in the ARA assay, suggest that the nitrogenase enzyme in ΔnifV induced nodules of these species is fully functional. However, the readily detected and proportionally increased amounts of “ethane” indicate that the nitrogenase enzyme complex (FeMo-cofactor) in nodules formed by ΔnifV mutant is different from the one in nodules induced by the WT strain. Moreover, the number of nodules formed by the ΔnifV mutant is double the amount formed by the WT strain and some of these ΔnifV induced nodules show signs of senescene. Thus, in contrast to the measured nitrogenase activity, all other observations with these three Aeschynomene species indicate that the symbiotic nitrogen fixation of the ΔnifV mutant is very inefficient. This difference can be explained by the fact that the nitrogenase activity as measured in the ARA assay (=reduction of only one of the triple bonds in acetylene) is not directly related to symbiotic nitrogen fixation which requires reduction of all triple bonds in dinitrogen. As shown in Fig. 1, in free-living conditions the nitrogenase activity of the ΔnifV mutant is not zero. Thus, when plants do not (or less rapid) sanction nodules that fix dinitrogen inefficiently, an increase in nodule number can give raise to a nitrogenase activity in the ARA assay which is close the one as observed for WT nodules. We hypothesize that the latter is the case in the three NF-independent Aeschynomene species showing a high nitrogenase activity with the ΔnifV mutant.
Fascinatingly, when the ΔnifV mutant is tested on the NF-dependent Aeschynomene species, A. afraspera, no effect of the mutation is detected indicating that the plant overcomes the absence of nifV in contrast to what is observed for A. evenia (Fig. 3). This difference between the two species is directly correlated with FEN1 expression. While two of the four FEN1 orthologs (FEN1-A1/A2) identified in A. afraspera genome (8x) are specifically expressed in the nodules (Fig. 5C and F), the only FEN1 ortholog identified in A. evenia (2x) displayed an opposite pattern of expression (down-expression in nodules) (Fig. 5B and D).
This non-requirement of bacterial NifV for symbiotic dinitrogen fixation is not a general feature of NF-dependent Aeschynomene species because in the second species tested, A. nilotica, we observed a drastic effect of the ΔnifV mutation on symbiotic efficiency (Fig. 4). The contrasting observations within the NF-dependent group render it difficult to propose a simple evolutionary scenario to explain the observed differences between Aeschynomene species. Nevertheless, knowledge acquired on the diversity and ecology of the Aeschynomene/Bradyrhizobium symbiosis could give some hints. It has been proposed that FEN1 was recruited from a housekeeping gene encoding an IPMS during the evolution of symbiosis5. The acquisition of this property, the specific control of FEN1 expression in the nodules, and a subsequent loss of nifV in the symbiont gives the host plant the capacity to control where and when the rhizobia make an active nitrogenase. Considering the high energy demand of the nitrogenase, this control represents a clear functional advantage. In nature, the NF-independent Aeschynomene species interact specifically with photosynthetic bradyrhizobia and these bacteria are known to nodulate only this group of plants. In addition, unlike other legumes, this group of Aeschynomene species form stem nodules. There are two possible explanations why the nifV genes are maintained in these bacteria. First, the photosynthetic properties of these bradyrhizobial strains make that in case of stem nodules they are less dependent on energy furnished by the plant for nitrogen fixation10. The need for the plant to control the energy consumption by the nodule tissue via homocitrate synthesis is thus less significant. Second, the ability of the bacteria to nodulate the stem, a surrounding very poor in nutrients makes the capacity to fix dinitrogen under free-living conditions an advantage for the bacterium as it will increase survival and infectivity. In the same vein, it is to highlight that A.caulinodans that also contains a nifV gene forms stems nodules on Sesbania rostrata.
In the group of NF-dependent Aeschynomene species, only a few members form stem nodules. In addition, the bacterial partner choice is less specific and symbiotic interactions with both photosynthetic and non-photosynthetic Bradyrhizobium strains are possible. Some of these non-photosynthetic strains (for example Bradyrhizobium USDA11017) do not contain a nifV gene. As NF-dependent Aeschynomene species show more heterogeneous symbiotic traits, the ability to overcome the lack of nifV in their bacterial partner may have a selective advantage.
Here, we show that in contrast to the model legume L. japonicus many Aeschynomene species do not supply homocitrate to the rhizobial partner during symbiosis. The subsequent question that arises is: Is Aeschynomene/photosynthetic Bradyrhizobium symbiosis an atypical example or are there other rhizobia/legume symbiosises that require rhizobial homocitrate synthesis to be efficient? As indicated above, the A. caulinodans/Sesbania rostrata symbiosis might be a second example, but what else? A survey of the genome sequences available show that ±10% of the sequenced rhizobia do contain a gene that is annotated as homocitrate synthase (Table S1). It would be very interesting to analyze if these rhizobia like photosynthetic bradyrhizobia are able to fix dinitrogen under free-living conditions and/or if the presence of the nifV gene is related to the absence of FEN1 expression by their natural host plant(s) like we observed for A. evenia. This knowledge will deepen our understanding on the evolution of the rhizobium / legume symbiosis and could contribute to a better selection of nitrogen fixing inoculum strains.
Methods
Bacterial strains and growth conditions
A detailed description of the construction of a nifV deletion strain can be found in the supplementary information section. Bradyrhizobium ORS285 and derivatives were grown in modified YM medium18 or BNM-B medium19. Escherichia coli strains were grown in Luria-Bertani medium (LB) at 37 °C. When required, the media were supplemented with kanamycin (100 μg/ml) or a mixture of kanamycin (120 μg/ml) and cefotaxime (20 μg/ml) for the selection of ORS285 clones in conjugation experiments.
Plant growth and acetylene reduction assay
A table of Aeschynomene species used in this study and their origin can be found in the Supplementary information section (Table S2). Sterilization of seeds, germination, plant growth and inoculation with bacterial strains were as described17. At the indicated times after inoculation as specified in the figure legends, photos of plants were taken, the number of nodules on the roots were counted and the acetylene reduction assay (ARA) was used to measure the nitrogenase enzyme activity17.
In vitro nitrogenase enzyme activity
Bacterial cultures were grown in liquid BNM-B medium containing 10 mM succinate under anoxic conditions and 10% acetylene. At the indicated times the amount of ethylene produced by the bacterial culture was measured by gas chromatography17. For complementation studies, homocitrate (Sigma-Aldrich; 10 mM final concentration) was added to the growth medium.
Transcriptome and real-time quantitative PCR expression analysis
Transcriptomic data for nodules of A. evenia (PI 225551) and A. afraspera (LSTM #1) were obtained and analysed as decribed15. Total RNA was extracted from lateral root regions (non-inoculated plant; ±1 cm around the exit of lateral roots) or nodules (plants inoculated with Bradyrhizobium ORS285) at 8 days after inoculation using the SV Total RNA Isolation system (Promega). Quantification of RNA, reverse transcription, real-time quantitative PCR and analysis of the data was performed as describe before20. Primers for quantitative PCR can be found in Table S3 the Supplementary information section.
Phylogenetic analysis
The procedure for the identification of FEN1 and IPMS orthologs in A. evenia (CIAT22838/PI 225551) and A. afraspera ((LSTM #1) is described in detail in the Supplementary information section. Using the obtained sequences a phylogenetic analysis was performed as described in16 and data are presented as rooted trees using the Cucumis IPMS as outgroup. GenBank/EMBL and Gene_ID numbers for sequences obtained and used for phylogenetic analysis can be found in Table S4 of the Supplementary information section. All DNA sequences generated in this study are deposited in Genbank under accession numbers KY412790–KY412799 and KY618805–KY618808.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the French National Research Agency, ANR-BugsInaCell-13-BSV7-0013 and ANR-AeschyNod-14-CE19-0005.
Author Contributions
N.N. and J.-F.A. conceived the experiment(s), N.N., J.-F.A., F.C., C.C. and D.G. conducted the experiment(s), C.K. delivered information, N.N., J.-F.A. and E.G. analyzed the result(s) and wrote the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00559-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cheng Q. Perspectives in biological nitrogen fixation research. J. Integr. Plant Biol. 2008;50:786–798. doi: 10.1111/j.1744-7909.2008.00700.x. [DOI] [PubMed] [Google Scholar]
- 2.Rubio LM, Ludden PW. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 2008;62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 3.Masson-Boivin C, Giraud E, Perret X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 2009;17:458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Hoover TR, Imperial J, Ludden PW, Shah VK. Homocitrate is a component of the iron-molybdenum cofactor of nitrogenase. Biochemistry. 1989;28:2768–2771. doi: 10.1021/bi00433a004. [DOI] [PubMed] [Google Scholar]
- 5.Hakoyama T, et al. Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature. 2009;462:514–517. doi: 10.1038/nature08594. [DOI] [PubMed] [Google Scholar]
- 6.Dreyfus BL, Elmerich C, Dommergues YR. Free-living Rhizobium strain able to grow on N(2) as the sole nitrogen source. Appl. Environ. Microbiol. 1983;45:711–713. doi: 10.1128/aem.45.2.711-713.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alazard D. Nitrogen fixation in pure culture by rhizobia isolated from stem nodules of tropical Aeschynomene species. FEMS Microbiol. Lett. 1990;68:177–182. doi: 10.1111/j.1574-6968.1990.tb04145.x. [DOI] [Google Scholar]
- 8.Hoover TR, Imperial J, Ludden PW, Shah VK. Homocitrate cures the NifV- phenotype in Klebsiella pneumoniae. J. Bacteriol. 1988;170:1978–1979. doi: 10.1128/jb.170.4.1978-1979.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott DJ, Dean DR, Newton WE. Nitrogenase-catalyzed ethane production and CO-sensitive hydrogen evolution from MoFe proteins having amino acid substitutions in an alpha-subunit FeMo cofactor-binding domain. J. Biol. Chem. 1992;267:20002–20010. [PubMed] [Google Scholar]
- 10.Giraud E, et al. Legumes symbioses: absence of nod genes in photosynthetic bradyrhizobia. Science. 2007;316:1307–1312. doi: 10.1126/science.1139548. [DOI] [PubMed] [Google Scholar]
- 11.Gourion B, et al. Bacterial RuBisCO is required for efficient Bradyrhizobium/Aeschynomene symbiosis. PLoS One. 2011;6:e21900. doi: 10.1371/journal.pone.0021900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortier V, Holsters M, Goormachtig S. Never too many? How legumes control nodule numbers. Plant. Cell Environ. 2012;35:245–258. doi: 10.1111/j.1365-3040.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 13.Chaintreuil C, et al. Evolution of symbiosis in the legume genus. Aeschynomene. New Phytol. 2013;200:1247–1259. doi: 10.1111/nph.12424. [DOI] [PubMed] [Google Scholar]
- 14.Kouchi H, Hata S. GmN56, a novel nodule-specific cDNA from soybean root nodules encodes a protein homologous to isopropylmalate synthase and homocitrate synthase. Mol. Plant. Microbe. Interact. 1995;8:172–176. doi: 10.1094/MPMI-8-0172. [DOI] [PubMed] [Google Scholar]
- 15.Chaintreuil C, et al. A gene-based map of the Nod factor-independent Aeschynomene evenia sheds new light on the evolution of nodulation and legume genomes. DNA Res. 2016;23:365–376. doi: 10.1093/dnares/dsw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaintreuil C, et al. The evolutionary dynamics of ancient and recent polyploidy in the African semiaquatic species of the legume genus. Aeschynomene. New Phytol. 2016;211:1077–1091. doi: 10.1111/nph.13956. [DOI] [PubMed] [Google Scholar]
- 17.Giraud E, Hannibal L, Fardoux J, Verméglio A, Dreyfus B. Effect of Bradyrhizobium photosynthesis on stem nodulation of Aeschynomene sensitiva. Proc. Natl. Acad. Sci. USA. 2000;97:14795–14800. doi: 10.1073/pnas.250484097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nouwen N, Fardoux J, Giraud E. NodD1 and NodD2 are not required for the symbiotic interaction of Bradyrhizobium ORS285 with Nod-factor-independent Aeschynomene legumes. PLoS One. 2016;11:805–811. doi: 10.1371/journal.pone.0157888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renier A, et al. Photosynthetic Bradyrhizobium sp. strain ORS285 synthesizes 2-O-methylfucosylated lipochitooligosaccharides for nod gene-dependent interaction with Aeschynomene plants. Mol. Plant. Microbe. Interact. 2011;24:1440–1447. doi: 10.1094/MPMI-05-11-0104. [DOI] [PubMed] [Google Scholar]
- 20.Czernic P, et al. Convergent evolution of endosymbiont differentiation in Dalbergioid and Inverted Repeat-Lacking Clade legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol. 2015;169:1254–1265. doi: 10.1104/pp.15.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


