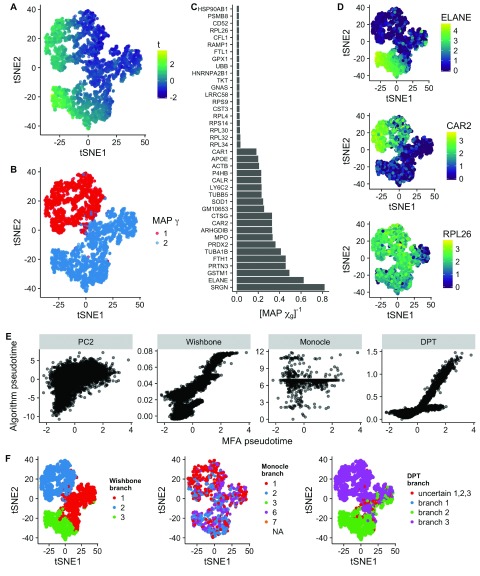
Figure 4. Inference of bifurcations in scRNA-seq data of 4,423 hematopoietic progenitor/stem cells differentiating into myeloid and erythroid precursors 3.
A tSNE representation colored by the maximum a posteriori (MAP) pseudotime. B Equivalent plot as A colored by MAP γ (branch assignment). C Inverse map χ showing both the 20 largest and 20 smallest values indicating which genes do and do not show differential behavior across the bifurcation. D tSNE representation of the dataset colored by gene expression. Both ELANE and CAR2 were predicted by the inverse χ values to show differing expression across the branches, while RPL26 was predicted to show similar expression. E Scatter plots of pseudotime values compared to those inferred by PC2, Wishbone, Monocle, and DPT. These had Pearson correlations of 0.54, 0.83, 0.01, and 0.78, respectively. F tSNE representations of the dataset colored by branch allocation of alternative algorithms shows good agreement with Wishbone and DPT.