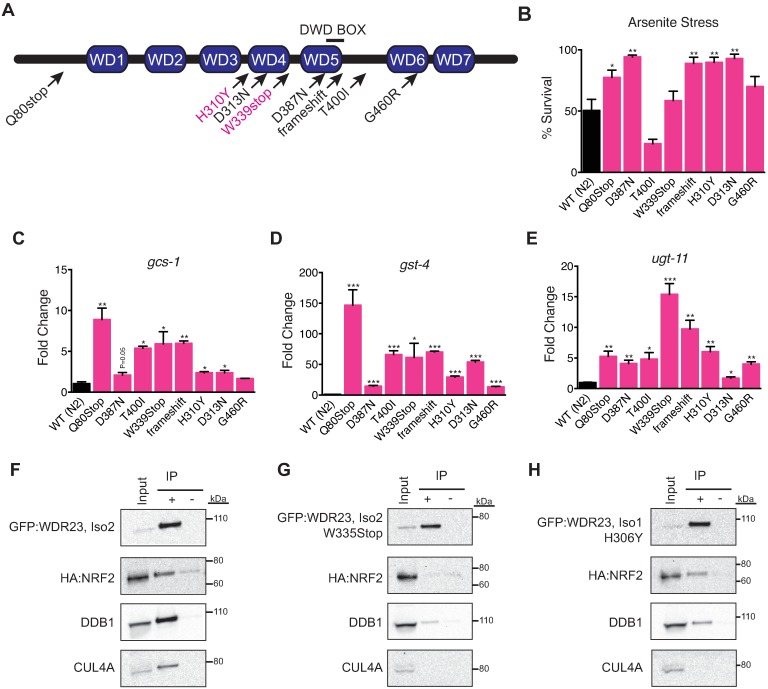
Fig 3. A central highly conserved domain of WDR23 facilitates binding to NRF2.
(A) Schematic of C. elegans WDR-23 and the identity of eight recessive loss of function alleles. Conserved residues between worm and human WDR23 that are mutated for structure function analysis are in pink. (B) Most mutations in WDR-23 result in an increase in cytoprotection from heavy metals. WT n = 6, Q80Stop n = 5, D387N n = 6, T400I n = 6, W339Stop n = 3, frameshift n = 6, H310Y n = 6, D313N n = 6, G460R n = 6. (C-E) Strains harboring mutant versions of WDR-23 display increased expression of the SKN-1/NRF2 cytoprotection targets gcs-1 (WT n = 3, Q80Stop n = 3, D387N n = 3, T400I n = 3, W339Stop n = 3, frameshift n = 3, H310Y n = 3, D313N n = 3, G460R n = 3) (C), gst-4 (WT n = 3, Q80Stop n = 3, D387N n = 3, T400I n = 3, W339Stop n = 3, frameshift n = 3, H310Y n = 3, D313N n = 3, G460R n = 3) (D), ugt-11 (WT n = 3, Q80Stop n = 3, D387N n = 3, T400I n = 3, W339Stop n = 3, frameshift n = 3, H310Y n = 3, D313N n = 3, G460R n = 3) (E). (F-H) As compared to cells over expressing wild type WDR23 and NRF2 (F), the WDR23(W335Stop) mutation (G) impairs binding of NRF2 and disrupts association with the DDB1-CUL4A complex, while the H306Y mutation modestly reduces binding of both (H). Data are mean ± s.e.m.; one-tailed t-test relative to control samples. *P<0.05,**P<0.01, ***P<0.001.