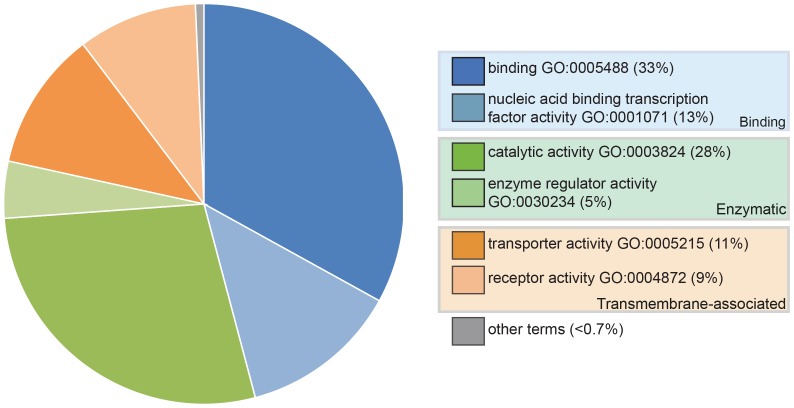
Figure 1. Pie chart showing the results of a PANTHER classification analysis by molecular function of 332 transcripts from the paddlefish lateral line-enriched dataset with associated gene ontology (GO) terms.
The percentage of function hits is indicated in parentheses. Binding activities GO:0005488 (blue) represent ~39% of the function hits (16% nucleic acid binding GO:0003637; 17% protein binding GO:0005515; 6% other binding, including calcium and lipid binding). Catalytic activities GO:0003824 (green) account for ~35% of the function hits (4% enzyme regulator activity GO:0030234; 9% transferase; and 11% each hydrolase and other catalytic activities). Transmembrane-associated activities (orange) represent ~19% of function hits (13% transporter GO:0005215; 6% receptor GO:0004872). The remaining ~7% of function hits are comprised of signal transducer GO:0004871, structural molecule GO:0005198 and antioxidant GO:0016209 activities.