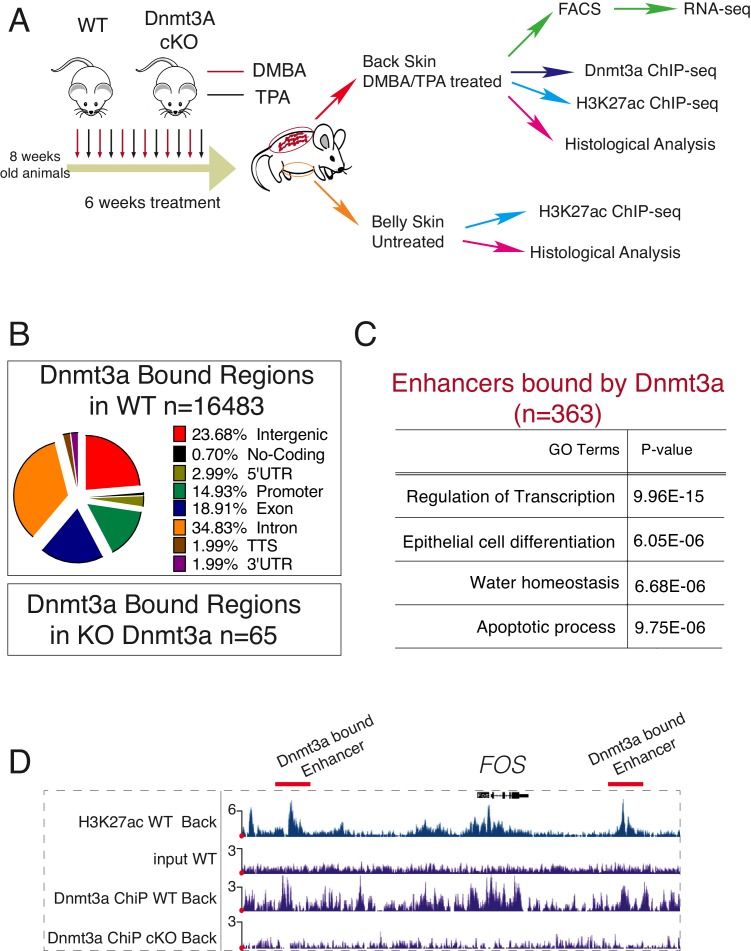
Figure 4. Dnmt3a binds a subset of enhancers in tumor cells.
(A) Schematic representation of a short treatment of DMBA/TPA in wild-type and Dnmt3a-cKO animals. (B) Genomic localizations of Dnmt3a determined by ChIP-seq of Dnmt3a in epidermal cells isolated from wild-type animals after 6 weeks of DMBA/TPA treatment. (C) Gene ontology analysis of the 363 H3K27ac-enriched regions (located at least 4 kb away from the TSS) also bound by Dnmt3a in isolated epidermis from wild-type animals after 6 weeks of DMBA/TPA. (D) Screenshot of enhancers bound by Dnmt3a in DMBA/TPA-treated skin in the FOS locus. All tracks are normalized to the number of mapped reads.