Figure 2.
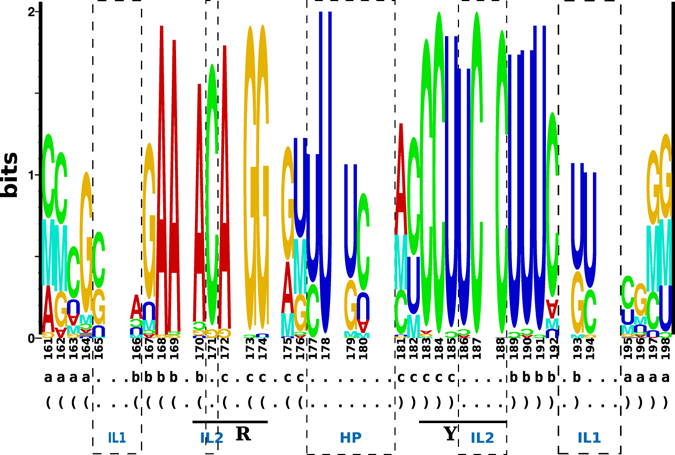
Structure logo of terminal right stem-loop. This logo46 is based on the same alignment (see Supplementary Fig. S2) of 95 pospiviroids as used for Fig. 1c. The height of nucleotide characters is proportional to the observed frequency of nucleotides in the alignment and the expected (a priori) frequency p = 0.25; nucleotide characters that appear less than expected are displayed up-side-down. The mutual information (marked by ‘M’) between base-paired positions, as indicated in the two bottom lines, is calculated as the relative entropy between the fractions of complementary bases and the number of basepairs one would expect by chance from the distribution of nucleotides at the involved positions. The nucleotide numbering is as in Fig. 1c; missing numbers denote that the respective column of the alignment contains mostly gaps. The RY motif critical for binding of Virp1 is outlined and marked by R and Y. The sequences of internal loops are marked by IL1 and IL2, respectively, and the hairpin loop sequence by HP.
