Figure 1.
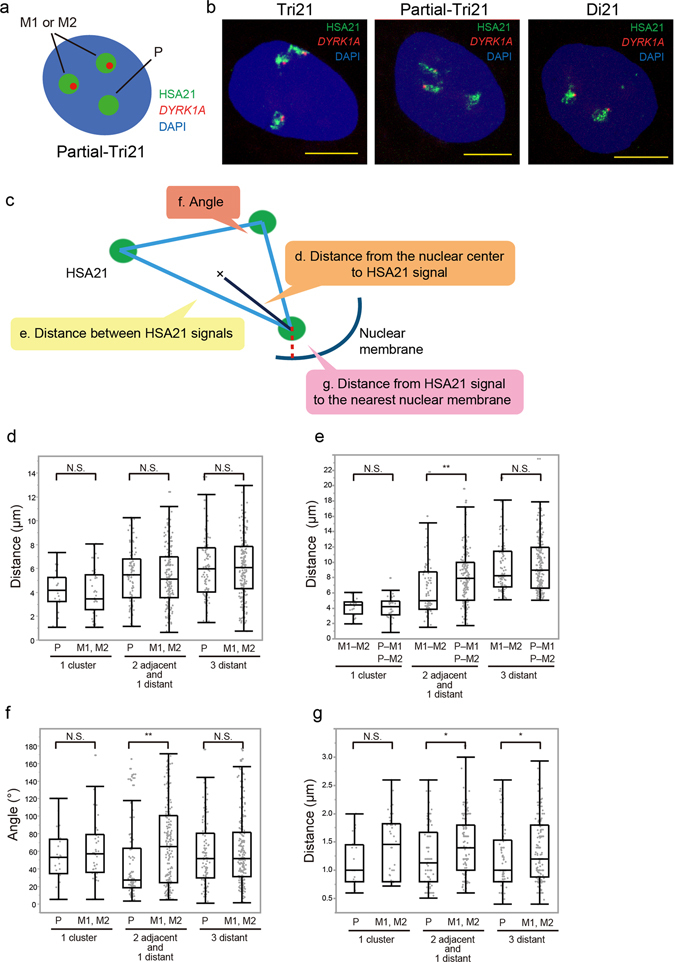
Chromosome positioning of paternal and maternal chromosome 21 in Partial-Tri21 iPSCs. (a) A schematic of the 3D-FISH analysis in Partial-Tri21 iPSCs. Signals of chromosome 21 (green) with or without a DYRK1A signal (red) represent chromosomes of maternal (M1/M2) or paternal origin, respectively. (b) Representative 3D-FISH images of Tri21, Partial-Tri21 and Di21 iPSCs. Chromosome 21 was labelled with Alexa488 (green), while the DYRK1A gene was labelled with Cy3 (red). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bars represent 10 μm. (c) A schematic representation of the distance measurements in 3D-FISH analysis. (d–g) Localisations of paternal and maternal chromosome 21 in Partial-Tri21 iPSCs. Localisation patterns of chromosome 21 categorised into three groups: 1 cluster (n = 20), 2 adjacent and 1 distant (n = 80), and all distant (n = 99). Measurements included (d) the distance from the nuclear centre to each copy of chromosome 21, (e) the distance between two copies of chromosome 21, (f) interior angles at the vertex and (g) the distance from chromosome 21 to the nearest nuclear membrane. The term “adjacent” here refers to the distance between copies of chromosome 21 being under 5 μm. Box plot represents 25th–75th percentile range ± min–max. p values were determined by the Mann–Whitney U test. *p < 0.05, **p < 0.01.
