Figure 1.
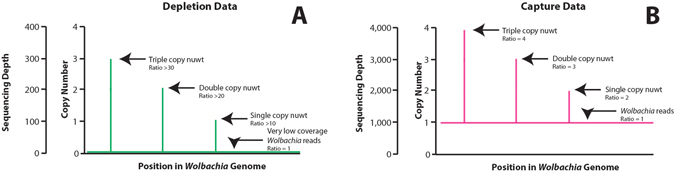
Schematic Depiction of Theoretical Differences Between the Capture and Depletion Data. This schematic depicts the differences between (A) a theoretical depletion data set and (B) a theoretical capture data set. The depletion data (A) is depicted here as low sequencing depth (<10X depth) on the Wolbachia genome and 100X depth on a typical single copy Brugia gene. Therefore, a single copy nuwt would have 100X depth while a nuwt with 2 or 3 copies would have 200X or 300X depth, respectively. The ratio of the depth for 1, 2, or 3 copy nuwts relative to Wolbachia endosymbiont DNA would be >10, >20, or >30 respectively. The capture data (B) is depicted here as having the same sequencing depth for the Wolbachia genome and a typical single copy Brugia gene (1000X depth). Therefore, a single copy nuwt would have 2000X depth while a nuwt with 2 or 3 copies would have 3000X or 400X depth, respectively. The ratio of the depth for 1, 2, or 3 copy nuwts relative to Wolbachia endosymbiont DNA would be 2, 3, or 4 respectively. This illustrates the effects of two important factors in the actual datasets examined in this study: (a) absolute sequencing depth (where capture depletion) and (b) the ratio of the sequencing depth for a Wolbachia sequence found as a nuwt relative to the same sequence found in the endosymbiont genome (where depletion capture).
