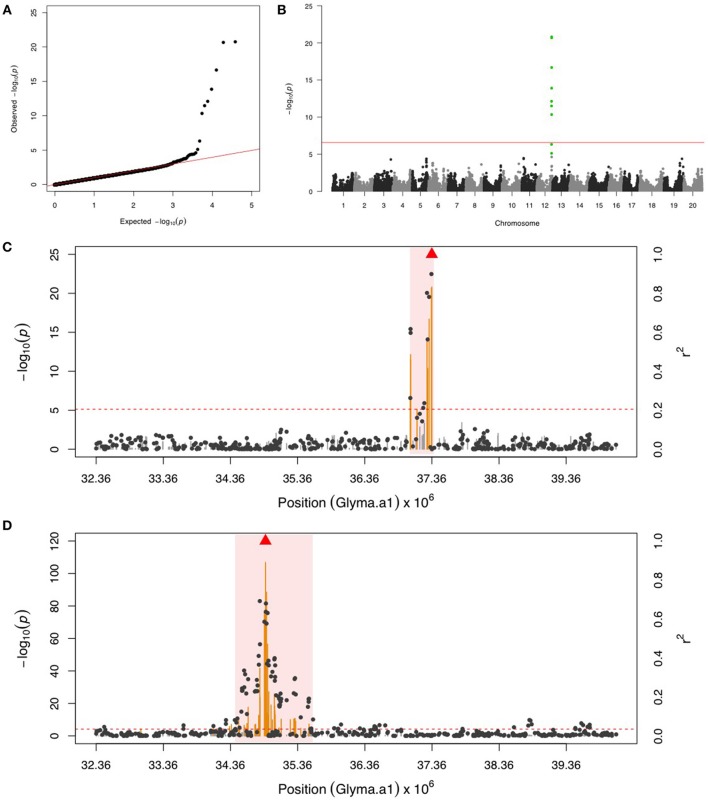
Figure 2.
GWAS for potato leafhopper (PLH) and pubescence density. (A) Quantile-quantile plot indicates the fitness of the regular MLM for PLH association analysis. (B) Manhattan plot identifies a significant locus for PLH resistance on chr 12. Green dots represent the nine SNPs with FDR below 0.05. Red horizontal line indicates the Bonferroni threshold. (C) Regional LD analysis for PLH locus that surrounds the most significant SNP (ss715612746), which is below the red triangle. The pairwise LD between SNPs (gray dots) to ss715612746 follows the right y axis. The orange and gray color for lines indicate SNPs with FDR below or above 0.05, respectively, following the left y axis. The red horizontal dash line represents the minimal significant level at the cutoff of FDR 0.05. The pink background highlights the high LD region where candidate genes were examined. (D) Regional LD analysis for pubescence density that surrounds the most significant SNP (ss715612489), shown in red triangle. The pairwise LD between SNPs (gray dots) to ss715612489 follows the right y axis. The orange and gray color for lines indicate SNPs with FDR below or above 0.05, respectively, following the left y axis. The red horizontal dashed line represents the minimal significant level at the cutoff of FDR 0.05. The pink background highlights the high LD region where candidate genes were examined. The genomic region for PLH resistance (C) and pubescence density (D) is exactly the same, but these two loci are not overlapping.