Figure 7.
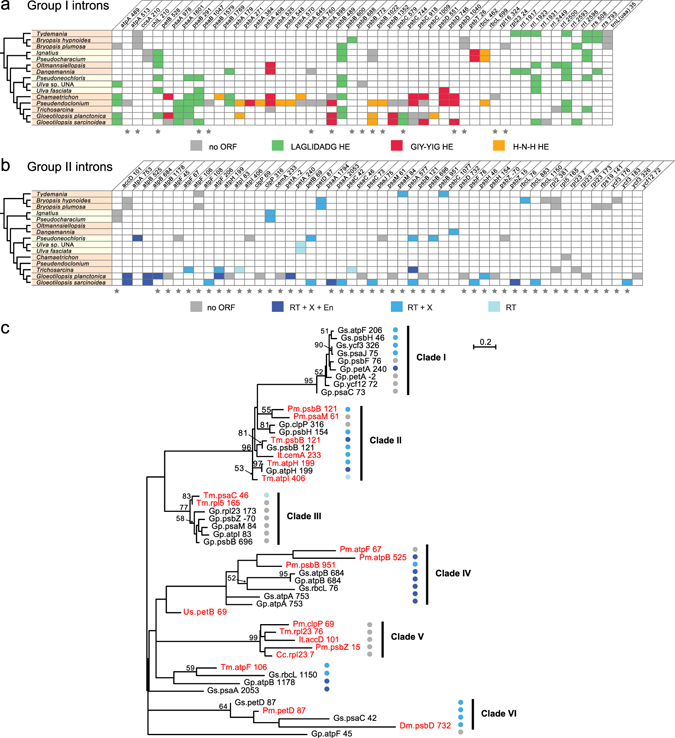
Introns in ulvophycean chloroplast genomes. (a) Distribution of group I introns. (b) Distribution of group II introns. A grey box denotes an intron lacking an ORF, whereas a colored box represents an intron containing an ORF (see the color code for the type of intron-encoded protein). Stars denote the intron insertion sites that have not been observed in other groups of chlorophytes. Intron insertion sites in protein-coding and tRNA genes are given relative to the corresponding genes in the Mesostigma chloroplast genome67; insertion sites in rrs and rrl are given relative to E. coli 16S and 23S rRNAs, respectively. For each insertion site, the position corresponding to the nucleotide immediately preceding the intron is reported. Abbreviations: EN, H-N-H endonuclease; RT, reverse transcriptase; X, intron maturase. (c) Phylogenetic relationships among group II introns of the Ignatiales, Oltmannsiellopsidales, Ulvales and Ulotrichales. The tree shown here was inferred by RAxML analysis of an alignment of 124 nucleotides corresponding to domains IA, IVB, V and VI of the core secondary structure. Bootstrap support values higher than 50% are reported on the nodes. Colored circles denote the types of intron ORF (see the color code on panel b). Note that clades I through IV were identified in a previous phylogenetic study of Gloeotilopsis group II introns31. Names of the taxa newly examined in the present investigation are indicated in red. Abbreviations: Cc, Chamaetrichon capsulatum; Dm, Dangemannia microcystis; Gp, Gloeotilopsis planctonica; Gs, Gloeotilopsis sarcinoidea; It, Ignatius tetrasporus; Pm, Pseudoneochloris marina; Tm, Trichosarcina mucosa; Us, Ulva sp. UNA00071828.
