Figure 1.
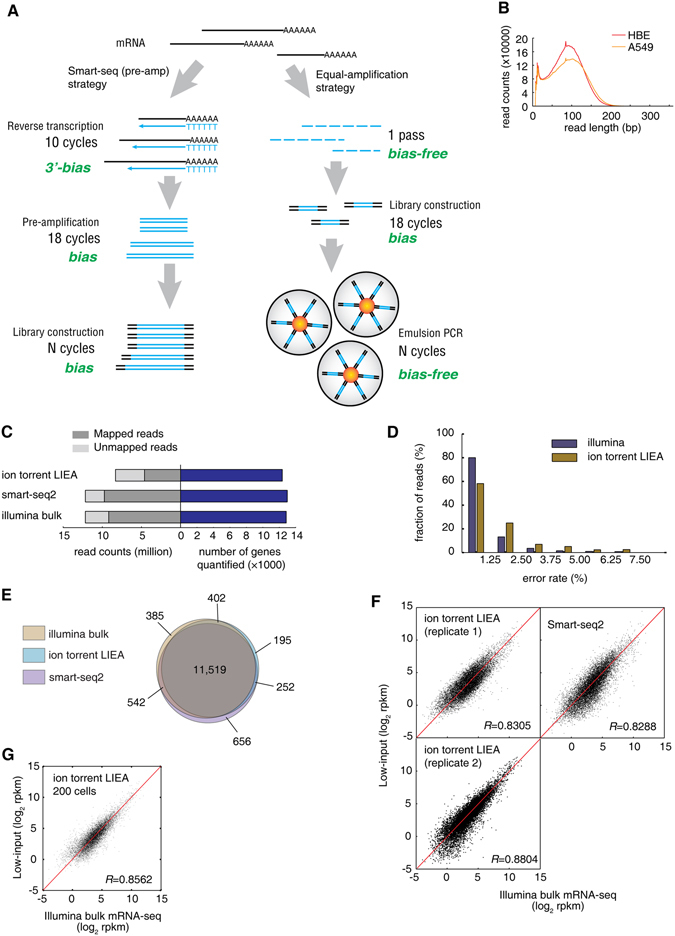
Low-input and low-bias ion torrent mRNA-seq. (A) Principle of the ion-torrent LIEA method and comparison to the smart-seq (pre-amplification) strategy. (B) Read length distribution of ion-torrent low-input mRNA-seq. (C) Comparison of the total read count, mapped read count and number of genes quantified by ion torrent LIEA method, smart-seq2 and Illumina bulk RNA-seq, for HBE cell line. (D) Error rate distribution of two platforms. An error is defined as a mismatch or indel nucleotide when aligned to the reference sequence. (E) Overlap of quantified genes of the three methods. (F) Comparison of the gene expression quantification using two low-input methods against Illumina bulk mRNA-seq, all for HBE cells. Ion torrent LIEA method was carried out twice as biological replicates. (G) Comparison of the gene expression quantification using LIEA method with 200 cells as starting material and Illumina bulk mRNA-seq.
