Figure 3.
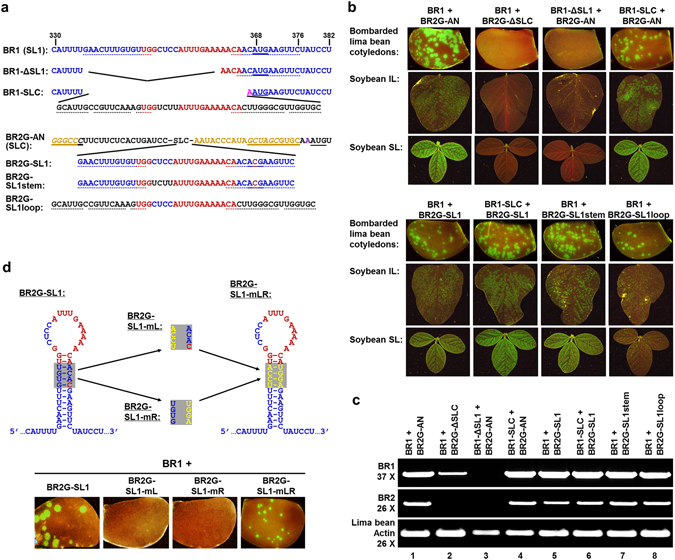
SL1 and SLC of BPMV are interchangeable. (a) Sequences of the SL1 and SLC portions of BR1 and BR2G-AN, respectively, and their deletion/replacement mutants. SL1 and SLC sequences are in blue and black letters, respectively; with the conserved loop nts in red. The nts involved in stems are underlined with dots. The AUG start codon in SL1, and its mutated form (ACG), are underlined with solid lines. Note that the last fifteen and nine nts of SL1 were retained in BR1-ΔSL1 and BR1-SLC, respectively, to preserve the AUG translational start codon. In BR2G derivatives containing SL1 segments, the AUG within the SL1 stem was changed to ACG to avoid its potential translational conflict with the RNA2 start codon downstream. (b) Lima bean cotyledons bombarded with various mutants, along with IL and SL images of soybean infected with the corresponding viruses. (c) RT-PCR detection of BR1 and BR2G RNAs in lima bean cotyledons bombarded with the mutants. (d) The stem of SL1 needs to remain base-paired in order for SL1 to be functional. Top: the predicted structure of SL1 in BR2G backbone, along with the three mutations (mL, MR, and mLR) introduced into the shaded area of the stem. Both mL and mR were predicted to disrupt the stem, whereas mLR should restore the base pairs with different sequences. Bottom: lima bean cotyledons bombarded with the constructs indicated above showing the restoration of replicability by the mLR mutant.
