Figure 3.
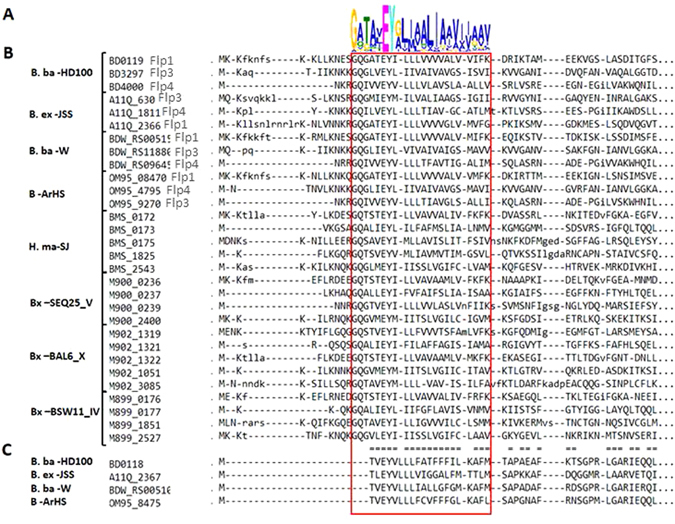
Multiple alignment of predicted flp sequences. Sequence logo representation representation70 of a Flp pilin sequence position weight matrix (PWM), depicting the canonic Flp pilin motif (GXXXXEY and a stretch of hydrophobic amino acids). (A) Multiple alignment of Flp pilin sequences identified by the PWM in various BALO’s with the flp pilin motif indicated by a red outline. (B) Flp2 sequences are truncated forms of Flp pilins, both the N terminus proximal glycine (G) and the amino acid that follows it are missing, creating motif: MXXEY and a stretch of hydrophobic amino acids. The protein logo was created with MEME60. Multiple alignments were calculated using GLAM260, 71. The Flp and the Flp2 multiple alignments were aligned to each other using the Compass program version 2.4572, with the similar regions indicated by the ‘=’ marks in between the multiple alignments. Lower case letters are regions not found to be significantly aligned by GLAM2. BALO designations are as in Fig. 2.
